Abstract
Gastrointestinal stromal tumors (GISTs) are KIT expressing spindle cell, epithelioid and rarely pleomorphic mesenchymal tumors. The majority of GISTs show gain-of-function KIT mutations. However, GISTs without KIT mutations and GISTs with weak or lack of immunohistochemical KIT expression have also been reported. Recently, gain-of-function mutations in exon 18 (activation loop) and exon 12 (juxtamembrane domain) of the PDGFRA were identified in such tumors. The purpose of this study was to test the hypothesis that PDGFRA mutation may define a specific clinicopathologic subgroup of GISTs. A total of 447 KIT exon 11 (juxtamembrane domain) mutation-negative GISTs were studied. DNA samples were obtained from formaldehyde-fixed paraffin-embedded tissues. Genomic sequences of PDGFRA exons 18 and 12 were evaluated for the mutations by PCR amplification and direct sequencing. PDGFRA exon 18 mutations were identified in 122 of 346 (35.3%) gastric GISTs and two of 75 (2.7%) intestinal GISTs. A great majority of these mutations represented simple T to A missense mutation at the codon 842 leading to substitution of the valine for aspartic acid (D842 V). However, in-frame deletions and deletions with point mutations clustering between codons 841–847 were found in approximately 23% of all exon 18 mutations. Mutations in PDGFRA exon 12 were found only in 10 of 170 (5.8%) gastric and one of 54 (1.9%) intestinal GISTs negative for KIT exon 11 and PDGFRA exon 18 mutations. There were seven substitutions of aspartic acid for valine at codon 561 (V561D) and four in-frame deletions with point mutations clustering between codons 566 and 571. The majority of GISTs with PDGFRA mutations had pure or predominant epithelioid morphology. Low mitotic activity, ≤5 mitoses/50HPF was detected in 81% of analyzed GISTs including larger, >5 cm tumors. Based on long-term follow-up (average 135 months), a majority (83.5%) of GISTs with PDGFRA mutations followed a benign course.
Similar content being viewed by others
Main
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor of the gastrointestinal tract and is probably related to interstitial cells of Cajal (ICC) or their progenitor cells. Morphologically, GISTs are cellular, spindle cell, epitheloid or rarely pleomorphic tumors with some site-related variation. A great majority of GISTs express KIT; however, in some cases KIT immunoreactivity is weak. Occasionally, KIT-negative GISTs have been reported.1
Gain-of-function KIT mutations in juxtamembrane (JM) domain (exon 11) have been commonly reported in GISTs.2, 3, 4 Subsequently, mutations in KIT extracellular (EC) domain (exon 9) and tyrosine kinase (TK) domains (exons 13 and 17) were found in KIT exon 11 mutation-negative tumors.5, 6, 7 Mutational alteration of KIT leads to ligand-independent activation (phosphorylation) of KIT TK and has transforming effects in vitro.2 Germline KIT mutations similar to those identified in sporadic GISTs have been reported in human familial GIST syndrome characterized by hyperpigmentation and/or urticaria pigmentosa, ICC hyperplasia, and GISTs.8, 9, 10, 11, 12, 13, 14 Introduction of a KIT exon 11 activating mutation, found in a case of human familial GIST syndrome, into the mouse genome reproduced features of human familial GIST syndrome in a mouse model and confirmed the critical role of KIT alteration in GIST development.15
More recently, platelet-derived growth factor receptor α (PDGFRA) was shown to be pathologically activated in GISTs. Gain-of-function PDGFRA mutations affecting the activation loop (exon 18 [TK II]) and JM domain (exon 12) were found in 14 of 40 (35%) KIT mutation-negative (KIT wild-type [WT]) GISTs. It was suggested that mutational activation of KIT or PDGFRA are mutually exclusive and represent two different alternative oncogenic events leading to similar biological consequences.16
KIT and PDGFRA display extensive structural homology and are members of the type III TK receptor family.17, 18 Activation of TK type III receptors by their ligands leads to downstream phosphorylation of substrate proteins and subsequently activates networks of signal transduction pathways that regulate important cell functions.19 Pathologic activation of TK receptors correlates with enhanced proliferation and development of cancer; its inhibition is considered an important therapeutic approach in oncology.20
Recently, imatinib mesylate (Gleevec/Glives, formerly STI571, Novartis, Switzerland) was successfully introduced in the treatment of clinically advanced, metastatic GISTs.21, 22 However, based on ‘in vitro’ studies, it has been suggested that KIT and PDGFRA activated by mutation in the TK II domain might not respond well to Gleevec-based TK- inhibition.23, 24, 25 Recent clinical studies on the response of metastatic GISTs to the Gleevec-based treatment revealed limited therapeutic effect in PDGFRA TK II mutant tumors; however, GISTs with PDGFRA JM mutations maintained a response similar to GISTs with KIT JM mutations.26
Assessment of the impact of new GIST therapy based on inhibition of KIT and PDGFRA TK activity requires delineation of the natural history of GISTs without treatment. Recent studies have been suggested that GISTs defined by certain type of KIT activating mutations might show different tumor natural history.27, 28, 29 The purpose of this study was to analyze a large series of GISTs defined by different types of PDGFRA mutations and delineate their clinicopathologic features and natural history.
Materials and methods
Tissue Material and Clinical Data
A total of 1000 GISTs were obtained from the files of the Armed Forces Institute of Pathology (AFIP), Washington, DC. Tumors were diagnosed using previously described morphologic and immunohistochemical criteria including evaluation of KIT (CD 117), CD34, α-smooth muscle actin (SMA), desmin (DES), and S100-protein.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 The analyzed tumors were divided into four histological categories, epithelioid (Ep), spindle cell (Sp) and tumors with mixed epithelioid and spindle cell features. The latter ‘mixed’ category was divided into subcategories, epithelioid with focal spindle cell component (Ep/sp) and spindle cell tumors with focal epithelioid component (Sp/ep). Mitoses were counted in 50 high-power fields (HPF), a total area of 5 mm2. Primary GISTs were distributed into six groups defining predicted clinical behavior based on tumor size and mitotic activity without knowledge of the outcome. Criteria used to classify tumor into clinicopathologic, prognostic groups were reported previously31 and are listed in Table 1. Clinical follow-up data were obtained and managed according to institutional review board approval. Patients treated with Gleevec were not included in this study as case accrual antedated availability of this drug.
Molecular Studies
DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissues. In total, 1000 GISTs were evaluated for KIT exon 11 mutations as previously described.3, 6 In addition, all nongastric GISTs without KIT exon 11 mutations were previously screened for KIT exon 9 mutations.29 We did not screen all tumors for mutations in KIT exons 13 and 17, because these mutations are very rare, each being seen in less than 1% of GISTs.6, 27, 32 However, six GISTs with known mutations in KIT exon 13 or 17 were excluded from the study.
KIT-WT exon 11 gastric GISTs and KIT-WT exon 11 and exon 9 nongastric GISTs (KIT-WT GISTs) were evaluated for mutations in PDGFRA exons 18 and 12 by PCR amplification and direct sequencing of PCR products. In addition, 60 randomly selected GISTs with documented KIT mutations in exon 11 (KIT-MT GISTs) were screened for mutations in PDGFRA exon 18, and 25 of them were also evaluated for mutations in PDGFRA exon 12. Primer sequences were those previously reported.16 Cycling conditions and the reaction mix were the standard ones as recommended by PE Applied Biosystem (Foster City, CA, USA). Annealing temperatures were 50 and 55°C, respectively, for the amplification of exons 18 and 12. In some cases, reading of forward sequencing data of PDGFRA exon 18 was affected by the presence of deletion/insertion single nucleotide polymorphism (SNP) in intron 17, listed in SNP database at http://www.ncbi.nlm.nih.gov under SNP ID: rs3830355. In such cases, PDGFRA exon 18 was sequenced a second time using forward primer P18.4F 5′AGTGTGTCCACCGTGATCTG 3′ located 3′ to the polymorphic site.
All PDGFRA mutation-positive GISTs (PDGFRA-MT GISTs) were divided into two groups: PDGFRA-MT exon 12 GISTs and PDGFRA-MT exon 18 GISTs. Clinicopathologic profiles of GISTs assigned to the different groups were examined.
Statistical Methods
The χ2-test, Kruskal–Wallis test and Wilcoxon rank-sum test were used for statistical analysis. Any P<0.05 was considered statistically significant.
Results
Types, Frequency and Distribution of PDGFRA Mutations among GISTs
In total, 447 KIT-WT tumors were found among 1000 well-documented GISTs and screened for mutations in PDGFRA exon 18. Subsequently, 242 KIT-WT and PDGFRA-WT exon 18 were evaluated for mutations in PDGFRA exon 12. In summary, PDGFRA mutations were identified in 139 cases. There were two types of PDGFRA mutations: point mutations (PM) and deletions sometimes coexisting with point mutations (DEL/DEL+PM). Genomic sequences and predicted sequences of mutant PDGFRA protein identified in this study are shown in Figure 1.
Mutations in PDGFRA exon 18 were found in 128 of 447 (28.6%) of the KIT-WT GISTs and were much more common than the ones affecting exon 12. A great majority (96%) of them represented T to A missense mutations leading to substitution of valine for aspartic acid at codon 842 (D842 V). However, in four cases variant missense mutations at codon 842 were identified with aspartic acid substituted by tyrosine (D842Y) or isoleucine (D842I). Substitution of tyrosine for aspartic acid was also found at codon 846 (D846Y) in one case. DEL/DEL+PM clustering between codons 841–847 represented 22.7% of PDGFRA exon 18 mutations. The most common mutations in this category were deletions IMHD843–846 and DIMH842–845 found in eight and six of 29 GISTs with DEL/DEL+PM, respectively.
PDGFRA exon 12 mutations were found in 11 of 242 (4.5%) KIT-WT and PDGFRA-WT exon 18 GISTs. There were seven A to T missense mutations resulting in substitution of aspartic acid for valine at codon 561 (V561D) and four almost identical DEL+PMs (SPDGHE566-571 K or SPDGHE566-571R) resulting in the loss of five PDGFRA amino-acid residue and subsequent substitution of one.
A great majority of PDGFRA exon 18 mutations, 122 of 128 (95%), were found in gastric tumors. Only two of 54 (3.7%) were identified in tumors of small intestinal origin. However, two primary GISTs diagnosed in the omentum and one in the retroperitoneum also had these mutations. PDGFRA exon 12 mutations were found in 10 of 170 (5.8%) gastric and one of 10 colonic GISTs and in none of 36 small intestinal GISTs. The frequency of PDGFRA exon 18 mutations among KIT-WT GISTs and PDGFRA exon 12 mutations among KIT-WT and PDGFRA-WT exon 18 GISTs is shown in Table 2.
In all, 60 and 25 GISTs with mutations in KIT exon 11 were screened for PDGFRA exons 18 and 12 mutations, respectively, and all revealed PDGFRA-WT sequences.
Demographic and Clinicopathologic Profile of PDGFRA-MT GISTs
There were 139 patients consisting of 86 male and 53 female patients. The overall male to female ratio was 1.6:1. Among the patients with PDGFRA-MT exon 18 GISTs, male predominated over female patients 87 to 47 (a 1.8:1 male-to-female ratio). In contrast, female patients predominated over men 4 to 7 (a 0.6:1 male-to-female ratio) among 11 patients with PDGFRA-MT exon 12 GISTs. However, the difference in sex distribution between cohorts defined by PDGFRA mutations was statistically nonsignificant (χ2-test, P=0.7).
The patient age ranged from 28 to 89 years with median age 64 years and average age 63 years, respectively. Although the patients' average age was 63–64 years in all subcohorts defined by different types of PDGFRA mutations, the average age of male patients was 7 years younger than that of female patients in the cohort defined by PDGFRA exon 18 mutation. Based on Wilcoxon rank-sum test, this difference was statistically significant (P=0.0014). The distribution of patient age and sex are shown in Table 3.
The tumor size of PDGFRA-MT GISTs ranged from 0.9 to 27 cm with an average maximum diameter of 7.3 cm. These cases were almost equally divided into tumor size categories ≤5 and >5 cm. In all, 20% of analyzed PDGFRA-MT GISTs were >10 cm and only two GISTs (1.5%) were ≤2 cm. However, GISTs with 842 PMs with average maximum diameter of 6.5 cm tended to be smaller than the ones with DEL/DEL+PM with average maximum diameter 9.5 cm. Based on Kruskal–Wallis test, this difference was statistically significant (P=0.017). The distribution of PDGFRA-MT GISTs in tumor size categories is shown in Table 4.
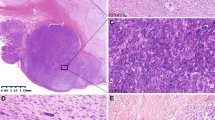
The majority of gastric PDGFRA-MT GISTs were histologically purely epithelioid or had an epithelioid component. Pure spindle cell morphology was seen only in 25 (19%) of 132 analyzed tumors. All three intestinal GISTs were spindle cell tumors, while all GISTs primary diagnosed in omentum and in the retroperitoneum had pure epithelioid morphology. Examples of purely epithelioid and spindle cell GISTs are shown in Figure 2. Distribution of PDGFRA-MT GISTs in the different morphological categories is shown in Table 5.
Examples of histology and KIT expression pattern and PDGFRA mutational status in three GISTs. Epithelioid GISTs (a) with diffuse, moderate KIT expression (b); spindle cell GIST (c) with diffuse, strong KIT expression (d); epithelioid GISTs diagnosed as primary in the omentum (e) with diffuse, moderate KIT expression (f); PDGFRA PM D842V (g) and D846Y (h) found in epithelioid GISTs (a, e); PDGFRA DEL+PM SPDGHE 566-571K (i) found in spindle cell GIST (c). Vertical arrows and horizontal arrow indicate point mutations and deletions, respectively.
A great majority of gastric PDGFRA-MT GISTs, 105 of 130 (81%) showed low mitotic activity (≤5 mitosis/50 HPF). Moreover, no mitotic activity (0/50 HPF) was found in 39% of these tumors. The average mitotic count was only 3/50 HPF. However, GISTs with DEL /DEL+PM in PDGFRA exon 18 showed two times higher mitotic activity compared to the tumors with PMs in codon 842. Based on the Kruskal–Wallis test, mitotic index was significantly associated with type of PDGFRA mutation (P=0.0045). Distribution of gastric PDGFRA-MT GISTs in the different categories of mitotic activity is shown in Table 6.
All three intestinal GISTs had ≤5 mitosis/50 HPF, whereas one GIST primary diagnosed in the omentum and one from the retroperitoneum had >5 mitosis/50 HPF.
KIT expression was immunohistochemically documented in 121 of 139 (87%) of PDGFRA-MT GISTs. In all, 108 tumors (78%) had at least 10% of KIT-positive tumor cells. In 13 cases, only scattered KIT-positive tumor cells were seen. In total, 18 PDGFRA-MT GISTs were KIT negative. Nine of these tumors expressed CD34. However, two cases had only scattered CD34-positive tumor cells. Seven cases showed focal (1–10%) expression of SMA. In four cases, none of the analyzed immunohistochemical markers (KIT, CD34, SMA, DES, and S100) was expressed. CD34 expression was seen in 98 of 136 (72%) PDGFRA-MT GISTs. A total of 86 tumors (66%) had at least 10% of CD34-positive tumor cells and 71 of them showed KIT coexpression in >10% of tumor cells. In all, 42 of 134 (31%) PDGFRA-MT GISTs expressed SMA but only 31 tumors in >10% of tumor cells. DES expression was detected in 10 of 110 (9%); however, in five cases, it was limited to scattered tumor cells. S100 protein was evaluated in 115 tumors and detected in less than 2% of analyzed cases. There was no correlation between immunohistochemical profile and type of PDGFRA mutation. Figure 2 shows the histology of three different GISTs, KIT expression in those tumors, and mutational status of PDGFRA.
Based on previously published criteria31 PDGFRA-MT GISTs were classified into six prognostic groups by tumor size and mitotic activity in order to evaluate the likelihood of GIST malignant behavior. In all, 42% of PDGFRA-MT GISTs were placed in the first or second prognostic groups characterized by small size ≤5 cm and mitotic activity not exceeding 5/50 HPF and considered to be of no or low malignant potential. In all, 44% were assigned to group 3 of uncertain malignant potential, characterized by low mitotic activity but >5 cm tumor size. Only 16% of analyzed tumors were assigned to groups 5 and 6 with size over 2 cm and mitotic rate >5/50 HPF indicating malignant potential. However, 27% of GISTs with DEL/DEL+PM, compared to only 11% of the ones with 842 PMs in exon 18 were assigned to groups 5 and 6. The distribution of PDGFRA-MT GISTs in six prognostic groups is shown in Table 7.
Clinical Behavior of PDGFRA-MT GISTs
Clinical follow-up data were available in 91 of 139 PDGFRA mutation-positive GISTs. In total, 41 (45.1%) patients were alive with no evidence of disease with average follow-up of 135 months (range 1–370), 24 (26.4%) died of unrelated causes, 16 (17.6%) died of unknown causes, and 10 (11.9%) showed evidence of progressive disease.
Seven of 10 patients with evidence of progressive disease died of the disease and three were alive with the disease at the end of follow-up. In three cases liver metastases or intra-abdominal dissemination were reported at the time of surgery and in two cases metastatic disease developed after 4 and 9 months of clinical follow-up. One patient died after 67 months, while the survival time of four others who died of tumor ranged from 107 to 214 months; the average was 165.5 months.
In all, 16 patients with incomplete follow-up data died of unknown causes. Survival time in these cases ranged from 4 to 245 months (average 72 months). However, more than 25% of these patients had a relatively long survival time ranging from 5 to 20 years. Based on tumor size, mitotic rate, and survival time, we estimated that five patients from this category could have developed progressive disease if complete data would have been available. This would have increased the frequency of tumors with malignant clinical course to 16.5%.
Discussion
In this study, we screened a large number of KIT-WT exon 11 GISTs from different locations for mutations in PDGFRA TK II and JM domains. We also evaluated the frequency of these mutations and established the clinicopathologic profile of PDGFRA-MT GISTs.
Gain-of-function PDGFRA mutations were recently found in GISTs and shown to be mutually exclusive with KIT mutations.16 These mutations represent a mechanism alternative to KIT mutation for pathologic activation of tyrosine kinase (TK) receptor signaling pathways.16, 25 Previously, KIT mutations were the only known type of TK activation in GISTs,2 and were even believed ubiquitous by some, prior to the discovery of PDGFRA mutations.7 Although more than 20 GISTs with PDGFRA mutations have been reported,16, 25, 26 little is known about clinicopathologic profile of PDGFRA-MT GISTs.
Here, we report genomic and predicted amino-acid sequences of 139 PDGFRA mutations found in GISTs and evaluate the clinicopathologic features of PDGFRA-MT GISTs. Based on our findings, the vast majority of these mutations cluster in TK II (exon 18) domain and were identified in 28.6% of exon 11 KIT-WT GISTs. One missense mutation, D842 V was particularly common and represented 73% of all PDGFRA TK II domain mutations. A similar high frequency (80%) of D842 V mutations was reported recently in the series of 10 exon 18 PDGFRA-MT GISTs.16 A D842 V PDGFRA mutation corresponds to the D816 V KIT mutation previously identified in mast cell tumors, cell lines thereof and germ cell tumors.33, 34, 35, 36
Two variant point mutations involving the same codon, D842Y and D842I identified in this study, were found in four cases only and have to be considered rare molecular events in GISTs. The only other isolated PDGFRA exon 18 PM was a substitution of tyrosine for the highly conserved aspartic acid at codon 846 (D846Y). The PDGFRA amino-acid residue at the position 846 is homologous to aspartic acid coded at the position 820 in KIT. The D820Y KIT mutation and more recently PDGFRA D846Y have been reported in familial GIST syndromes.13, 37 In our study, D846Y mutation was found in one epithelioid GISTs primary diagnosed in the omentum. Although normal control tissue was not available for molecular studies to exclude the possibility of germline mutation, the clinical data did not support familial GIST syndrome in this case.
In all, 13 different PDGFRA exon 18 DEL/DEL+PMs were identified in this study. Review of previously published four PDGFRA exon 18 DEL/DEL+PMs revealed two more deletions, DEL HDSN845–848 and DEL I843.16, 26 Although PDGFRA DEL/DEL+PMs appeared to be heterogeneous, all of them clustered in the region of eight PDGFRA amino-acid residues, 841–848. Since this region harbors also all PDGFRA PMs, it should be considered as a major PDGFRA mutational ‘hot spot’ for GISTs.
PDGFRA JM domain (exon 12) mutations were found only in 4.5% of exon 11 KIT-WT GISTs and were six times less frequent than mutations in TK II domain as previously reported.16 In this study, the most common PDGFRA exon 12 mutation was a V561D missense mutation, identified in seven of 11 (63.7%) of exon 12 PDGFRA-MT GISTs. Review of seven exon 12 PDGFRA-MT GISTs reported in the literature revealed similar 57.1% frequency of this mutation.16, 25, 26 PDGFRA V561D mutation corresponds to V559D KIT mutation reported in sporadic human GISTs.2
The remaining four exon 12 PDGFRA-MT GISTs had DEL+PM SPDGHE566-577 K (n=3) or its variant SPDGHE566-577R (n=1). The latter was reported once (16). Review of 11 PDGFRA JM domain mutations from this study and seven others reported in the literature shows that all of them clustered between 560 and 577 PDGFRA amino-acid residues and this region should be considered as a minor mutational ‘hot spot’ in GISTs.
In this study, PDGFRA mutations had a clear predilection to gastric over intestinal GISTs, as 95% of these tumors were gastric and only three of them (2%) were of intestinal origin. PDGFRA mutations were also associated with epithelioid tumor morphology, low mitotic rate, and relatively low overall mortality. Associations between specific type of KIT mutation and clinicopathologic profile of GISTs have been recently reported. Gastric GISTs with internal tandem duplications in 3′ KIT JM domain were associated with spindle cell morphology, relatively large tumor size and low mitotic activity. In contrast, tumors with insertion of AY residues at codon 502 in KIT EC domain showed a predilection to intestinal location and malignant clinical behavior.27, 28, 29
Some of the early studies reported that KIT exon 11 mutations (mostly DEL/DEL+PMs) are more common in large and malignant GISTs3, 4 and suggested adverse prognostic significance of such mutations.3, 4, 38 However, others39 have shown KIT mutations in diminutive, clinically indolent incidental tumors. Subsequently, KIT PMs were suggested to be linked to less malignant clinical behavior.40 Larger numbers of cases with specific types of mutations have to be analyzed in comparison to confirm the specific trends in tumor behavior.
In this study, gastric GISTs with PDGFRA DEL/DEL+PMs had larger size and a higher mitotic activity than tumors with PDGFRA PMs. Moreover, tumors from this category were more frequently assigned to group 5 and 6 of predicted malignant clinical behavior than GISTs with PDGFRA PMs. Follow-up studies based on a larger number of GISTs with DEL/DEL+PMs will be necessary to confirm the adverse prognostic significance of this type of PDGFRA mutation.
PDGFRA expression was reported in the substantial number of KIT-WT and KIT-MT GISTs25, 26 with the tendency to be stronger in KIT-WT GISTs.25 In this study, we did not report immunohistochemical expression of PDGFRA because commercially available anti-PDGFRA antibodies lacked specific reactivity in FFPE tissues (data not shown). We have evaluated immunohistochemically PDGFRA-MT GISTs for KIT expression and found that the majority of these tumors expressed KIT. However, in 31 cases (22.3%) only scattered KIT-positive cells or no KIT expression was seen. Although KIT expression and KIT tyrosine phosphorylation was previously reported in four KIT-WT GISTs,7 mutually exclusive expression of KIT and PDGFRA was recently demonstrated, respectively in a few KIT-MT and PDGFRA-MT GISTs.16 However, recently Hirota et al. suggested that constitutively activated mutant PDGFRA binds and activates the WT-KIT in ‘in vitro’ transfection systems and in ‘in vivo’ tumor tissue as well25 opening the possibility of convergence of PDGFRA and KIT pathways. Extensive RNA/protein studies on FFPE and fresh/frozen tissues employing a spectrum of techniques and based on a significant number of PDGFRA-MT GISTs are necessary to establish the expression profiles of the PDGFRA-MT GISTs.
Based on this study on PDGFRA mutations and our recent studies on KIT mutations in GISTs,28, 29 some types of KIT and PDGFRA mutations appear to be nonrandomly distributed among GISTs from different sites and with different clinicopathologic parameters. This may be one of the factors leading to apparent discrepancies in the frequency of KIT mutations in GISTs reported by previously published studies.4, 6, 7, 26, 27, 38, 39, 41, 42, 43 A lower frequency of KIT mutations in some of these studies is probably related to a higher number of KIT-WT, PDGFRA-MT gastric GISTs. Similarly, the apparent higher frequencies of AY insertions at codon 502 in KIT EC domain might have resulted from selection favoring malignant intestinal tumors GISTs.26, 29
In summary, we have shown that GISTs defined by PDGFRA TK II (exon 18) mutations represent a subset of tumors almost exclusively occurring in the gastric location. Histologically, these tumors often show epithelioid morphology and usually have low mitotic activity and most follow a benign course. Therefore, detection of PDGFRA mutation could be an additional feature that identifies gastric GISTs with a high probability of benign behavior. However, full understanding of the biological differences between these and other gastric GISTs will require additional clinical and biological studies.
References
Miettinen M, Lasota J . Gastrointestinal stromal tumors (GISTs): definition, occurrence, pathology, differential diagnosis and molecular genetics. Pol J Pathol 2003; 54:3–24.
Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998;279:577–580.
Lasota J, Jasinski M, Sarlomo-Rikala M, et al. Mutations in exon 11 of c-kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am J Pathol 1999;154:53–60.
Taniguchi M, Nishida T, Hirota S, et al. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res 1999;59:4297–4300.
Lux ML, Rubin BP, Biase TL, et al. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol 2000;156:791–795.
Lasota J, Wozniak A, Sarlomo-Rikala M, et al. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. A study of two hundred cases. Am J Pathol 2000;157:1091–1095.
Rubin BP, Singer S, Tsao C, et al. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res 2001;61:8118–8121.
Nishida T, Hirota S, Taniguchi M, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet 1998;19:323–324.
Hirota S, Okazaki T, Kitamura Y, et al. Cause of familial and multiple gastrointestinal autonomic nerve tumors with hyperplasia of interstitial cells of Cajal is germline mutation of the c-kit gene. Am J Surg Pathol 2000;24:326–327.
Beghini A, Tibiletti MG, Roversi G, et al. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer 2001;92:657–662.
Isozaki K, Terris B, Belghiti J, et al. Germline-activating mutation in the kinase domain on KIT gene in familial gastrointestinal stromal tumors. Am J Pathol 2001; 157:1581–1585.
Maeyama H, Hidaka E, Ota H, et al. Familial gastrointestinal stromal tumor with hyperpigmentation: association with a germline mutation of the c-kit gene. Gastroenterology 2001;120:210–215.
Hirota S, Nishida T, Isozaki K, et al. Familial gastrointestinal stromal tumors associated with dysphagia and novel type germline mutation of KIT gene. Gastroenterology 2002;122:1493–1499.
Robson ME, Glogowski E, Sommer G, et al. Pleomorphic characteristic of a germ-line KIT mutation in a large kindred with gastrointestinal stromal tumors, hyperpigmentation, and dysphagia. Clin Cancer Res 2004;10:1250–1254.
Sommer G, Agosti V, Ehlers I, et al. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the KIT receptor tyrosine kinase. Proc Natl Acad Sci USA 2003;100:6706–6711.
Heinrich MC, Corless CL, Duensing A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science 2003;299:708–710.
Yarden Y, Kuang WJ, Yang Feng T, et al. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J 1987;6:3341–3351.
Matsui T, Heidaran M, Miki T, et al. Isolation of a novel receptor cDNA establishes the existence of two PDGF receptor genes. Science 1989;243:800–804.
Pawson T . Regulation and targets of receptor tyrosine kinases. Eur J Cancer 2002;38 (Suppl 5):S3–S10.
Levitzki A . Tyrosine kinases as targets for cancer therapy. Eur J Cancer 2002;38 (Suppl 5):S11–S18.
van Oosterom AT, Judson I, Verweij J, et al. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet 2001;358:1421–1423.
Demetri GD, von Mehren M, Blanke CD, et al. Efficacy and saafety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 2002;347:472–480.
Longley BJ, Reguera MJ, Ma Y . Classes of c-KIT activating mutations: proposed mechanisms of action and implications in disease classification and therapy. Leuk Res 2001;25:571–576.
Chen H, Isozaki K, Kinoshita K, et al. Imatinib inhibits various types of activating mutant kit found in gastrointestinal stromal tumors. Int J Cancer 2003; 105:130–135.
Hirota S, Ohashi A, Nishida T, et al. Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors. Gastroenterology 2003;125:660–667.
Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003;23:4342–4349.
Antonescu CR, Tschernyavsky SK, Sommers G, et al. Association of KIT exon 9 mutations with non-gastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin Cancer Res 2003;15:3329–3337.
Lasota J, Dansonka-Mieszkowska A, Stachura T, et al. Gatrointestinal stromal tumors (GIST) with internal tandem duplications in 3' end of KIT juxtamembrane domain occur predominantly in stomach and generally seem to have a favorable course. Mod Pathol 2003;16:1257–1264.
Lasota J, Kopczynski J, Sarlomo-Rikala M, et al. KIT 1530ins6 mutation defines subset of predominantly malignant gastrointestinal stromal tumors (GIST) of intestinal origin. Hum Pathol 2003;34:1306–1312.
Miettinen M, Sobin LH, Sarlomo-Rikala M . Immunohistochemical spectrum of gastrointestinal stromal tumors at different sites and their differential diagnosis with other tumors with a special reference to CD117 (KIT). Mod Pathol 2000;13:1134–1142.
Miettinen M, El-Rifai W, Sobin LH, et al. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors—a review. Hum Pathol 2002;33:466–477.
Kinoshita K, Isozaki K, Hirota S, et al. c-kit Gene mutation at exon 17 or 13 is very rare in sporadic gastrointestinal stromal tumors. J Gastroenterol Hepatol 2003;18:147–151.
Furitsu T, Tsujimura T, Tono T, et al. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest 1993;92:1736–1744.
Nagata H, Worobec AS, Oh CK, et al. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci USA 1995;92:10560–10564.
Longley BJ, Tyrrell L, Lu S-Z, et al. Somatic c-KIT activating mutation in urticaria pigmentosa and aggressive mastocytosis: establishement of clonality in a human mast cell neoplasm. Nat Genet 1996;12:312–314.
Tian Q, Frierson HF Jr, Krystal GW, et al. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol 1999;154:1643–1647.
Chompret A, Kannengiesser C, Barrois M, et al. PDGFRA germline mutation in a family with multiple cases of gastrointestinal stromal tumor. Gastroenterology 2004;126:318–321.
Ernst SI, Hubbs AE, Przygodzki RM, et al. KIT mutation portends poor prognosis in gastrointestinal stromal/smooth muscle tumors. Lab Invest 1998;78: 1633–1636.
Corless CL, McGreevey L, Haley A, et al. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol 2002;160:1567–1572.
Singer S, Rubin BP, Lux ML, et al. Prognostic value of KIT mutation type, mitotic activity, and histological subtype in gastrointestinal stromal tumors. J Clin Oncol 2002;20:3898–3905.
Moskaluk CA, Tian Q, Marshall CR, et al. Mutations of c-kit JM domain are found in a minority of human gastrointestinal stromal tumors. Oncogene 1999;18:1897–1902.
Sakurai S, Fukasawa T, Chong JM, et al. c-kit gene abnormalities in gastrointestinal stromal tumors (tumors of intestitial cells of Cajal). Jpn J Cancer Res 1999;90:1321–1328.
Wardelmann E, Neidt I, Bierhoff E, et al. c-kit Mutations in gastrointestinal stromal tumors occur preferentially in the spindle rather than in the epithelioid cell variant. Mod Pathol 2002;15:125–136.
Acknowledgements
The opinions and assertions contained herein are the expressed views of the authors and are not to be construed as official or reflecting the views of the Departments of the Army or Defense. The authors thank Ms Virginia Achstetter and Ms Wanda King for skillful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
A Dansonka-Mieszkowska was research fellow at the Department of Soft Tissue Pathology, AFIP, Washington, DC, USA (2002–2003).
Rights and permissions
About this article
Cite this article
Lasota, J., Dansonka-Mieszkowska, A., Sobin, L. et al. A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Lab Invest 84, 874–883 (2004). https://doi.org/10.1038/labinvest.3700122
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3700122
Keywords
This article is cited by
-
Identification of an Activating PDGFRA Deletion in a Novel Sinonasal Soft Tissue Neoplasm
Head and Neck Pathology (2023)
-
Unique gastrointestinal stromal tumor with PDGFRA D842Y mutation—evaluation of in vivo sensitivity to imatinib
memo - Magazine of European Medical Oncology (2021)
-
Gastrointestinal stromal tumor: a review of current and emerging therapies
Cancer and Metastasis Reviews (2021)
-
Nationwide evaluation of mutation-tailored treatment of gastrointestinal stromal tumors in daily clinical practice
Gastric Cancer (2021)
-
Pretreatment Tumor DNA Sequencing of KIT and PDGFRA in Endosonography-Guided Biopsies Optimizes the Preoperative Management of Gastrointestinal Stromal Tumors
Molecular Diagnosis & Therapy (2020)