Abstract
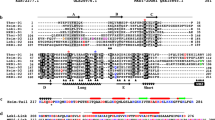
It was thought until recently that bacteria lack the actin or tubulin filament networks that organize eukaryotic cytoplasm. However, we show here that the bacterial MreB protein assembles into filaments with a subunit repeat similar to that of F-actin—the physiological polymer of eukaryotic actin. By elucidating the MreB crystal structure we demonstrate that MreB and actin are very similar in three dimensions. Moreover, the crystals contain protofilaments, allowing visualization of actin-like strands at atomic resolution. The structure of the MreB protofilament is in remarkably good agreement with the model for F-actin, showing that the proteins assemble in identical orientations. The actin-like properties of MreB explain the finding that MreB forms large fibrous spirals under the cell membrane of rod-shaped cells, where they are involved in cell-shape determination. Thus, prokaryotes are now known to possess homologues both of tubulin, namely FtsZ, and of actin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Schmidt, A. & Hall, M. N. Signaling to the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14, 305–338 (1998).
Straub, F. B. Actin. Stud. Inst. Med. Chem. Univ. Szeged 2, 3–15 (1942).
Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. Atomic model of the actin filament. Nature 347, 44–49 (1990).
Egelman, E. H., Francis, N. & DeRosier, D. J. F-actin is a helix with a random variable twist. Nature 298, 131–135 (1982).
Steinmetz, M. O. et al. An atomic model of crystalline actin tubes: combining electron microscopy with X-ray crystallography. J. Mol. Biol. 278, 703–711 (1998).
Aebi, U., Fowler, W. E., Isenberg, G., Pollard, T. D. & Smith, P. R. Crystalline actin sheets—their structure and polymorphism. J. Cell Biol. 91, 340–351 (1981).
Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. Atomic-structure of the actin–DNase-I complex. Nature 347, 37–44 (1990).
McLaughlin, P. J., Gooch, J. T., Mannherz, H. G. & Weeds, A. G. Structure of gelsolin segment-1-actin complex and the mechanism of filament severing. Nature 364, 685–692 (1993).
Schutt, C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C. W. & Lindberg, U. The structure of crystalline profilin β-actin. Nature 365, 810–816 (1993).
Chik, J. K., Lindberg, U. & Schutt, C. E. The structure of an open state of β-actin at 2.65 angstrom resolution. J. Mol. Biol. 263, 607–623 (1996).
Bork, P., Sander, C. & Valencia, A. An ATPase domain common to prokaryotic cell-cycle proteins, sugar kinases, actin, and Hsp70 heat-shock proteins. Proc. Natl Acad. Sci. USA 89, 7290–7294 (1992).
Kabsch, W. & Holmes, K. C. Protein motifs 2. The actin fold. FASEB J. 9, 167–174 (1995).
Flaherty, K. M., Delucaflaherty, C. & McKay, D. B. Three-dimensional structure of the ATPase fragment of a 70kDa heat-shock cognate protein. Nature 346, 623–628 (1990).
van den Ent, F. & Löwe, J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19, 5300–5307 (2000).
Hurley, J. H. et al. Structure of the regulatory complex of Escherichi coli Iii(Glc) with glycerol kinase. Science 259, 673–677 (1993).
Anderson, C. M., McDonald, R. C. & Steitz, T. A. Sequencing a protein by X-ray crystallography. Interpretation of yeast hexokinase B at 2.5 Å resolution by model building. J. Mol. Biol. 123, 1–13 (1978).
Wachi, M. et al. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169, 4935–4940 (1987).
Doi, M. et al. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J. Bacteriol. 170, 4619–4624 (1988).
Levin, P. A., Margolis, P. S., Setlow, P., Losick, R. & Sun, D. X. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174, 6717–6728 (1992).
Jones, L. J. F., Carballido-Lopez, R. & Errington, J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104, 913–922 (2001).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Flaherty, K. M., McKay, D. B., Kabsch, W. & Holmes, K. C. Similarity of the 3-dimensional structures of actin and the ATPase fragment of a 70-kDa heat-shock cognate protein. Proc. Natl Acad. Sci. USA 88, 5041–5045 (1991).
Lorenz, M., Popp, D. & Holmes, K. C. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J. Mol. Biol. 234, 826–836 (1993).
Tirion, M. M., Benavraham, D., Lorenz, M. & Holmes, K. C. Normal-modes as refinement parameters for the F-actin model. Biophys. J. 68, 5–12 (1995).
Page, R., Lindberg, U. & Schutt, C. E. Domain motions in actin. J. Mol. Biol. 280, 463–474 (1998).
Nogales, E., Downing, K. H., Amos, L. A. & Löwe, J. Tubulin and FtsZ form a distinct family of GTPases. Nature Struct. Biol. 5, 451–458 (1998).
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996).
Crowther, R. A., Henderson, R. & Smith, J. M. MRC image processing programs. J. Struct. Biol. 116, 9–16 (1996).
van den Ent, F., Lockhart, A., Kendrick-Jones, J. & Löwe, J. Crystal structure of the N-terminal domain of MukB: a protein involved in chromosome partitioning. Struct. Fold. Des. 7, 1181–1187 (1999).
Leslie, A. G. W. Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data (SERC Laboratory, Daresbury, UK, 1991).
Collaborative Computing Project No. 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Terwilliger, T. C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Terwilliger, T. C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000).
Turk, D. Weiterentwicklung eines Programms für Molekülgrafik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. Thesis, Technische Univ. München (1992).
Brünger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Engh, R. A. & Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A 47, 392–400 (1991).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Kraulis, P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Bacon, D. J. Raster 3D: photorealistic molecular graphics. Methods Enzymol. 277, 505–524 (1997).
Barton, G. J. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 6, 37–40 (1993).
Acknowledgements
We would like to thank S. Munro for critically reading the manuscript and the staff at ID14-4 of ESRF (Grenoble, France) for assistance with MAD data collection.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
van den Ent, F., Amos, L. & Löwe, J. Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 (2001). https://doi.org/10.1038/35092500
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35092500
This article is cited by
-
Actin cytoskeleton and complex cell architecture in an Asgard archaeon
Nature (2023)
-
Tubulin Cytoskeleton in Neurodegenerative Diseases–not Only Primary Tubulinopathies
Cellular and Molecular Neurobiology (2023)
-
Extracting phylogenetic dimensions of coevolution reveals hidden functional signals
Scientific Reports (2022)
-
Polymerization of Bacillus subtilis MreB on a lipid membrane reveals lateral co-polymerization of MreB paralogs and strong effects of cations on filament formation
BMC Molecular and Cell Biology (2020)
-
Chiral twisting in a bacterial cytoskeletal polymer affects filament size and orientation
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.