Abstract
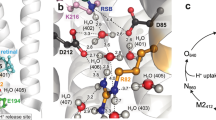
Bacteriorhodopsin, a membrane protein with a relative molecular mass of 27,000, is a light driven pump which transports protons across the cell membrane of the halophilic organism Halobacterium salinarum. The chromophore retinal is covalently attached to the protein via a protonated Schiff base. Upon illumination, retinal is isomerized. The Schiff base then releases a proton to the extracellular medium, and is subsequently reprotonated from the cytoplasm. An atomic model for bacteriorhodopsin was first determined by Henderson et al1, and has been confirmed and extended by work in a number of laboratories in the last few years2. Here we present an atomic model for structural changes involved in the vectorial, light-driven transport of protons by bacteriorhodopsin. A ‘switch’ mechanism ensures the vectorial nature of pumping. First, retinal unbends, triggered by loss of the Schiff base proton, and second, a protein conformational change occurs. This conformational change, which we have determined by electron crystallography at atomic (3.2 Å in-plane and 3.6 Å vertical) resolution, is largely localized to helices F and G, and provides an ‘opening’ of the protein to protons on the cytoplasmic side of the membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Henderson, R. et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213, 899–929 (1990).
Subramaniam, S. The structure of bacteriorhodopsin: an emerging consensus. Curr. Opin. Struct. Biol. 9, 462–468 (1999).
Subramaniam, S. et al. Protein conformational changes in the bacteriorhodopsin photocycle. J. Mol. Biol. 287, 145– 161 (1999).
Vonck, J. Structure of the bacteriorhodopsin mutant F219L N intermediate revealed by electron crystallography. EMBO J. 19, 2152 –2160 (2000).
Luecke, H., Schobert, B., Richter, H. -T., Cartailler, J. -P., & Lanyi, J. K. Structural changes in bacteriorhodopsin during ion transport at 2 Å resolution. Science 286, 255–260 ( 1999).
Sass, H.-J. et al. Structural alterations for proton translocation in the M state of wild-type bacteriorhodopsin. Nature 406, 649–653 (2000).
Dencher, N. A., Dresselhaus, D., Zaccai, G. & Büldt, G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc. Natl Acad. Sci. USA 86 , 7876–7879 (1989).
Koch, M. H. J. et al. Time-resolved X-ray diffraction study of structural changes associated with the photocycle of bacteriorhodopsin. EMBO J. 10, 521–526 (1991).
Kamikubo, H. et al. The last phase of the reprotonation switch in bacteriorhodopsin: The transition between the M-type and the N-type protein conformation depends on hydration. Biochemistry 36, 12282– 12287 (1996).
Subramaniam, S., Gerstein, M., Oesterhelt, D. & Henderson, R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 12, 1–8 (1993).
Oka, T. et al. Conformational change of helix G in the bacteriorhodopsin photocycle: Investigation with heavy atom labeling and X-ray diffraction. Biophys. J. 76, 1018–1023 (1999).
Fodor, S. P. et al. Chromophore structure in bacteriorhodopsin's N intermediate: implications for the proton pumping mechanism. Biochemistry 27, 7097–7101 (1988).
Elia, G. R., Childs, R. F., Britten, J. F., Yang, D. S. C. & Santarsiero, B. D. Structure and wavelength modification in retinylidene iminium salts. Can. J. Chem. 74, 591–601 (1996).
Santarsiero, B. D., James, M. N. G., Mahendran, M. & Childs, R. F. The crystal structure of N-methyl-N-phenyl-retinylideneiminium perchlorate: a structural model for the bacteriorhodopsin chromophore. J. Am. Chem. Soc. 112, 9416–9418 (1990).
Hamanaka, T., Mitsui, T., Ashida, T. & Kakudo, M. The crystal structure of all-trans retinal1. Acta Crystallogr. B 28, 214–222 ( 1972).
Stam, C. H. The crystal structure of a monoclinic modification and the refinement of a triclinic modification of vitamin A acid (retinoic acid), C20H 28O2. Acta Crystallogr. B 28, 2936–2945 (1972).
Simmons, C. J., Liu, R. S. H., Denny, M. & Seff, K. The crystal structure of 13-cis-retinal. The molecular structures of its 6-s-cis and 6-s-trans conformers. Acta Crystallogr. B 37 , 2197–2205 (1981).
Simmons, C. J., Asato, A. E. & Liu, R. S. H. Structure of All-trans-3,4-didehydroretinal (retinal2). Acta Crystallogr. C 42, 711–715 (1986).
Moltke, S. et al. The angles between the C-1-, C-5-, and C-9-methyl bonds of the retinylidene chromophore and the membrane normal increase in the M-intermediate of bacteriorhodopsin: Direct determination with solid-state H-2 NMR. Biochemistry 38, 11762–11772 (1999).
Griffiths, J. M. et al. Structural investigation of the active site in bacteriorhodopsin: Geometric constraints on the roles of Asp-85 and Asp-212 in the proton pumping mechanism from solid-state NMR. Biochemistry 39, 362–371 (2000).
Brown, L. S. et al. A local electrostatic change is the cause of the large scale protein conformation shift in bacteriorhodopsin. Proc. Natl Acad. Sci. USA 94, 5040–5044 ( 1997).
Schobert, B. & Lanyi, J. K. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257, 306– 313 (1982).
Hoff, W. D., Jung, K. H. and Spudich, J. L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Ann. Rev. Biophys. Biomol. Struct. 26, 223–258 (1997).
Altenbach, C. et al. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: a site-directed spin-labeling study. Biochemistry 35, 12470– 12478 (1996).
Faruqi, A. R., Henderson, R. & Subramaniam, S. Cooled CCD detector with tapered fibre optics for recording electron diffraction patterns. Ultramicroscopy 75, 235–250 (1999).
Grigorieff, N., Ceska, T. A., Downing, K. H., Baldwin, J. M. & Henderson, R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259, 393–421 (1996).
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Essen, L. O., Siegert, R., Lehmannn, W. D. & Oesterhelt, D. Lipid patches in membrane protein oligomers: Crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl Acad. Sci. USA 95, 11673 –1167 (1998).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Subramaniam, S., Henderson, R. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin . Nature 406, 653–657 (2000). https://doi.org/10.1038/35020614
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35020614
This article is cited by
-
Bioinspired supramolecular macrocycle hybrid membranes with enhanced proton conductivity
Nano Research (2024)
-
Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting
Nature Methods (2023)
-
True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins
Nature Structural & Molecular Biology (2022)
-
Collective exchange processes reveal an active site proton cage in bacteriorhodopsin
Communications Biology (2020)
-
Cryo-EM structure of human rhodopsin bound to an inhibitory G protein
Nature (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.