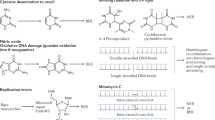
Abstract
The bacterial SOS response to unusual levels of DNA damage has been recognized and studied for several decades. Pathways for re-establishing inactivated replication forks under normal growth conditions have received far less attention. In bacteria growing aerobically in the absence of SOS-inducing conditions, many replication forks encounter DNA damage, leading to inactivation. The pathways for fork reactivation involve the homologous recombination systems, are nonmutagenic, and integrate almost every aspect of DNA metabolism. On a frequency-of-use basis, these pathways represent the main function of bacterial DNA recombination systems, as well as the main function of a number of other enzymatic systems that are associated with replication and site-specific recombination.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hanawalt, P. C. The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem. Photobiol. 5, 1– 12 (1966).
Skalka, A. in Mechanisms in Recombination (ed. Grell, R. F.) 421– 432 (Plenum, New York, 1974).
West, S. C., Cassuto, E. & Howard-Flanders, P. Mechanism of E. coli RecA protein directed strand exchanges in post-replication repair of DNA. Nature 294, 659–662 (1981).
Luder, A. & Mosig, G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: priming by RNA polymerase and by recombination. Proc. Natl Acad. Sci. USA 79, 1101–1105 (1982).
Formosa, T. & Alberts, B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47, 793– 806 (1986).
Zavitz, K. H. & Marians, K. J. Dissecting the functional role of PriA protein-catalysed primosome assembly in Escherichia coli DNA replication. Mol. Microbiol. 5, 2869– 2873 (1991).
Kuzminov, A. Collapse and repair of replication forks in Escheria coli. Mol. Microbiol. 16, 373–384 (1995).
Kogoma, T. Recombination by replication. Cell 85, 625 –627 (1996).
Sandler, S. J., Samra, H. S. & Clark, A. J. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics 143, 5–13 ( 1996).
Kogoma, T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61, 212–238 (1997).
McGlynn, P., Al, D. A., Liu, J., Marians, K. J. & Lloyd, R. G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol. 270, 212–221 (1997).
Cox, M. M. Recombinational crossroads—eukaryotic enzymes and the limits of bacterial precedents. Proc. Natl Acad. Sci. USA 94, 11764–11766 (1997).
Michel, B., Ehrlich, S. D. & Uzest, M. DNA double-strand breaks caused by replication arrest. EMBO J. 16, 430–438 (1997).
Seigneur, M., Bidnenko, V., Ehrlich, S. D. & Michel, B. RuvAB acts at arrested replication forks. Cell 95, 419–430 (1998).
Steiner, W. W. & Kuempel, P. L. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J. Bacteriol. 180, 6269–6275 (1998).
Mosig, G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 32, 379– 413 (1998).
Neilson, L., Blakely, G. & Sherratt, D. J. Site-specific recombination at dif by Haemophilus influenzae XerC. Mol. Microbiol. 31, 915–926 (1999).
Rangarajan, S., Woodgate, R. & Goodman, M. F. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli . Proc. Natl Acad. Sci. USA 96, 9224 –9229 (1999).
Bidnenko, V. et al. sbcS sbcC null mutations allow RecF-mediated repair of arrested replication forks in rep recBC mutants. Mol. Microbiol. 33, 846–857 (1999).
Saveson, C. J. & Lovett, S. T. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics 152, 5– 13 (1999).
McGlynn, P. & Lloyd, R. G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 27, 3049–3056 (1999).
Marians, K. J. PriA: at the crossroads of DNA replication and recombination. Prog. Nucleic Acids Res. Mol. Biol. 63, 39– 47 (1999).
Cox, M. M. Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acids Res. Mol. Biol. 63, 310– 366 (1999).
Kuzminov, A. Recombinatorial repair in Escherichia coli. Microbiol. Mol. Biol. Rev. 63, 751–813 ( 1999).
Arai, K. et al. Enzyme studies of phi X174 DNA replication. Prog. Nucleic Acid Res. Mol. Biol. 26, 9– 32 (1981).
Marians, K. J. Enzymology of DNA in replication in prokaryotes. CRC Crit. Rev. Biochem. 17, 153–215 ( 1984).
Clark, A. J. & Sandler, S. J. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 20, 125–142 (1994).
Clark, A. J. & Margulies, A. D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc. Natl Acad. Sci. USA 53, 451– 459 (1965).
Howard-Flanders, P. & Theriot, L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics 53, 1137–1150 (1966).
Nurse, P., Zavitz, K. H. & Marians, K. J. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J. Bacteriol. 173, 6686–6693 ( 1991).
Kuzminov, A. Instability of inhibited replication forks in E. coli. Bioessays 17, 733–741 ( 1995).
Liu, J. & Marians, K. J. PriA-directed assembly of a primosome on D loop DNA. J. Biol. Chem. 274, 25033 –25041 (1999).
Liu, J. I., Xu, L. W., Sandler, S. J. & Marians, K. J. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl Acad. Sci. USA 96, 3552 –3555 (1999).
Sandler, S. J. et al. dnaC mutations suppress defects in DNA replication and recombination associated functions in priB and priC double mutants in E. coli K-12. Mol. Microbiol. 34, 91–101 (1999).
Sandler, S. J. & Marians, K. J. Role of PriA in replication fork reactivation in Escherichia coli. J. Bacteriol. 182, 9–13 ( 2000).
Cox, M. M. A broadening view of recombinatorial DNA repair in bacteria. Genes to Cells 3, 65–78 ( 1998).
Steiner, W. W. & Kuempel, P. L. Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli. Mol. Microbiol. 27, 257– 268 (1998).
Skarstad, K. & Boye, E. Degradation of individual chromosomes in recA mutants of Escherichia coli. J. Bacteriol. 75, 5505–5509 ( 1993).
Marians, K. J. Prokaryotic DNA replication. Annu. Rev. Biochem. 61 , 673–719 (1992).
Anderson, D. G. & Kowalczykowski, S. C. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90, 77–86 (1997).
Courcelle, J., Carswell-Crumpton, C. & Hanawalt, P. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl Acad. Sci. USA 94, 3714–3719 (1997).
Kuzminov, A. & Stahl, F. W. Double-strand end repair via the RecBC pathway in Escherichia coli primes DNA replication. Genes Dev. 13, 345–356 ( 1999).
Shinagaw, H. SOS response as an adaptive response to DNA damage in prokaryotes. Exs 77, 221–235 ( 1996).
Tang, M. et al. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′ 2C mutagenic complex and RecA protein. Proc. Natl Acad. Sci. USA 95, 9755–9760 ( 1998).
Tang, M. J. et al. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc. Natl Acad. Sci. USA 96, 8919–8924 (1999).
Wagner, J. et al. The dinB gene encodes a novel E-coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell. 4, 281–286 (1999).
Woodgate, R. & Ennis, D. G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol. Gen. Genet. 229, 10–16 (1991).
Kaguni, J. M. & Kornberg, A. Replication initiated at the origin (oriC) of the E. coli chromosome reconstituted with purified enzymes. Cell 38, 183–190 (1984).
Kogoma, T., Cadwell, G. W., Barnard, K. G. & Asai, T. The DNA replication priming protein, PriA, is required for homologous recombination and double-strand break repair. J. Bacteriol. 178, 1258–1264 (1996).
Steiner, W., Liu, G., Donachie, W. D. & Kuempel, P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol. Microbiol. 31, 579–583 (1999).
Recchia, G. D., Aroyo, M., Wolf, D., Blakely, G. & Sherratt, D. J. FtsK-dependent and -independent pathways of Xer site-specific recombination. EMBO J. 18, 5724– 5734 (1999).
Galitski, T. & Roth, J. R. Pathways for homologous recombination between chromosomal direct repeats in Salmonella typhimurium. Genetics 146, 751–767 ( 1997).
Haber, J. E. DNA recombination: the replication connection. Trends Biochem. Sci. 24, 271–275 ( 1999).
Malkova, A., Ivanov, E. L. & Haber, J. E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA 93, 7131– 7136 (1996).
Morrow, D. M., Connelly, C. & Hieter, P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147, 371–382 ( 1997).
Lundblad, V. & Blackburn, E. H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73, 347–360 (1993).
Le, S., Moore, J. K., Haber, J. E. & Greider, C. W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 152, 143–152 (1999).
Griffith, J. D. et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 ( 1999).
Bosco, G. & Haber, J. E. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics 150, 1037–1047 ( 1998).
Chen, C., Umezu, K. & Kolodner, R. D. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2, 9– 22 (1998).
Cordeiro-Stone, M., Makhov, A. M., Zaritskaya, L. S. & Griffith, J. D. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 289, 1207–1218 (1999).
Opperman, T., Murli, S., Smith, B. T. & Walker, G. C. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc. Natl Acad. Sci. USA 96, 9218–9223 (1999).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cox, M., Goodman, M., Kreuzer, K. et al. The importance of repairing stalled replication forks. Nature 404, 37–41 (2000). https://doi.org/10.1038/35003501
Issue Date:
DOI: https://doi.org/10.1038/35003501
This article is cited by
-
A novel targeted NGS panel identifies numerous homologous recombination deficiency (HRD)-associated gene mutations in addition to known BRCA mutations
Diagnostic Pathology (2024)
-
Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli
Nature Communications (2023)
-
Single-molecule imaging of LexA degradation in Escherichia coli elucidates regulatory mechanisms and heterogeneity of the SOS response
Nature Microbiology (2021)
-
Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures
Nature Communications (2021)
-
Whole-genome sequencing and genomic-based acid tolerance mechanisms of Lactobacillus delbrueckii subsp. bulgaricus LJJ
Applied Microbiology and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.