Abstract
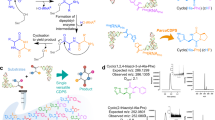
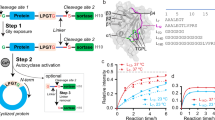
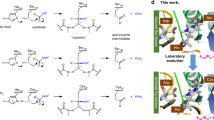
Designing an enzyme requires, among a number of parameters, the appropriate positioning of catalytic machinery within a substrate-binding cleft. Using the structures of cyclophilin–peptide complexes1,2,3,4, we have engineered a new catalytic activity into an Escherichia coli cyclophilin by mutating three amino acids, close to the peptide binding cleft, to form a catalytic triad similar to that found in serine proteases. In conjunction with cyclophilin's specificity for proline-bearing peptides, this creates a unique endopeptidase, cyproase 1, which cleaves peptides on the amino-side of proline residues. When acting on an Ala-Pro dipeptide, cyproase 1 has an efficiency (kcat/Km) of 0.7 × 104 M−1 s−1 and enhances the rate of reaction (kcat/kuncat) 8× 108-fold. This activity depends upon a deprotonated histidine and is inhibited by nucleophile-specific reagents, as occurs in natural serine proteases. Cyproase 1 can hydrolyse a protein substrate with a proline-specific endoprotease activity.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Clubb, R. T., Ferguson, S. B., Walsh, C. T. & Wagner, G. Three-dimensional solution structure of Escherichia coli periplasmic cyclophilin. Biochemistry 33, 2761–2772 (1994).
Konno, M., Ito, M., Hayano, T. & Takahashi, N. The substrate-binding site in Escherichia coli cyclophilin A preferably recognizes a cis-proline isomer or a highly distorted form of the trans-isomer. J. Mol. Biol. 256, 897–908 (1996).
Zhao, Y. & Ke, H. Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes. Biochemistry 35, 7362–7362 (1996).
Zhao, Y. & Ke, H. Crystal structure implies that cyclophilin predominantly catalyzes the trans to cis isomerization. Biochemistry 35, 7356–7362 (1996).
Kirby, A. J. Enzyme mechanisms, models, and mimics. Angew. Chem. Int. Ed. Engl. 35, 707–724 (1996).
Matthews, B. W., Craik, C. S. & Neurath, H. Can small cyclic peptides have the activity and specificity of proteolytic enzymes. Proc. Natl Acad. Sci. USA 91, 4103–4105 (1994).
Fersht, A. Enzyme Structure and Mechanism (W. H. Freeman, New York, (1985)).
Fischer, G., Wittmann-Liebold, B., Lang, K., Kiefhaber, T. & Schmid, F. X. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337, 476–478 (1989).
Galat, A. & Metcalfe, S. M. Peptidylproline cis/trans isomerases. Prog. Biophys. Mol. Biol. 63, 69–119 (1995).
Carter, P. & Wells, J. A. Dissecting the catalytic triad of a serine protease. Nature 332, 564–568 (1988).
Bode, W. & Huber, R. Natural protein proteinase inhibitors and their interaction with proteinase. Eur. J. Biochem. 204, 433–451 (1992).
Gamble, T. R. et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87, 1285–1294 (1996).
Kahne, D. & Still, W. C. Hydrolysis of a peptide bond in neutral water. J. Am. Chem. Soc. 110, 7529–7534 (1988).
Radzicka, A. & Wolfenden, R. Aproficient enzyme. Science 267, 90–93 (1995).
Stein, R. L. Mechanism of enzymatic and nonenzymatic prolyl cis-trans isomerization. Adv. Prot. Chem. 44, 1–24 (1993).
Maillère, B. et al. Immunogenicity of a disulphide-containing neurotoxin: presentation to T-cells requires a reduction step. Toxicon 33, 475–482 (1995).
Ménez, A., Montenay-Garestier, T., Fromageot, P. & Hélène, C. Conformation of two homologous neurotoxins. Fluorescence and circular dichroism studies. Biochemistry 19, 5202–5208 (1980).
Strynadka, N. C. J. et al. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature 359, 700–705 (1992).
Duggleby, H. J. et al. Penicillin acylase has a single-amino-acid catalytic centre. Nature 373, 264–268 (1995).
Scott, D. L. et al. Interfacial catalysis: the mechanism of phospholipase A2. Nature 250, 1541–1546 (1990).
Liu, J. L. & Walsh, C. T. Peptidyl-prolyl cis-trans isomerase from Escherichia coli: A periplasmic homolog of cyclophilin that is not inhibited by cyclosporin A. Proc. Natl Acad. Sci. USA 87, 4028–4032 (1990).
Plummer, T. H. J & Kimmel, M. T. An improved spectrophotometric assay for human carboxypeptidase N1. Anal. Biochem. 108, 348–353 (1980).
Kraulis, P. Molscript: a program to produce both detailed and schematic plots of protein structures. J.Appl. Crystallogr. 24, 946–950 (1991).
Merritt, E. A. & Murphy, M. Raster3D version 2.0 — a program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994).
Segel, I. H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems (John Wiley, New York, (1993)).
Acknowledgements
We thank J. Janin, F. Lederer and G. Robillard for useful and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Quéméneur, E., Moutiez, M., Charbonnier, JB. et al. Engineering cyclophilin into a proline-specific endopeptidase. Nature 391, 301–304 (1998). https://doi.org/10.1038/34687
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/34687
This article is cited by
-
What history tells us XLII. A ‘new’ view of proteins
Journal of Biosciences (2017)
-
Protein functional-group 3D motif and its applications
Chinese Science Bulletin (2000)
-
Protective effect of cyclosporin A and FK506 from nitric oxide‐dependent apoptosis in activated macrophages
British Journal of Pharmacology (1999)
-
Creation of a ribonuclease abzyme through site-directed mutagenesis
Nature Biotechnology (1998)
-
Erratum: Engineering cyclophilin into a proline-specific endopeptidase
Nature (1998)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.