Abstract
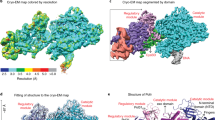
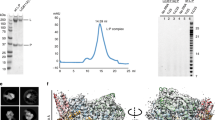
DURING transcription in E. coli, the DNA-dependent RNA polymerase locates specific promoter sequences in the DNA template, melts a small region containing the transcription start site, initiates RNA synthesis, processively elongates the transcript, and finally terminates and releases the RNA product. Each step is regulated by interactions between the polymerase, the DNA, the nascent RNA, and a variety of regulatory proteins and ligands1-3. The E. coli enzyme contains a catalytic core of two α-subunits, one β-and one β′-subunit, with relative molecular masses (Mr) of 36,512, 150,619 and 155,162, respectively2. The holoenzyme has an additional regulatory subunit, normally σ-70, of Mr 70,236. Preparations may also contain the ω-subunit (Mr∼10,000), which can be removed without affecting any known properties of the enzyme2. Because the amino-acid sequences of the β- and β′-subunits are homologous to those of the largest subunits of the yeast, Drosophila and murine RNA polymerases4-7, it seems likely that essential features of the three-dimensional structure and catalytic mechanism of RNA polymerase are also conserved across species. Crystals of RNA polymerase suitable for X-ray analysis have not yet been obtained, but two-dimensional crystals of E. coli RNA polymerase holoenzyme can be grown on positively charged lipid layers8. Electron microscopy of these crystals in negative stain shows the enzyme in projection as an irregularly shaped complex ∼100 x 100 x 160 Å in size. We have now determined the three-dimensional structure by electron microscopy of negatively stained, two-dimensional crystals tilted at various angles to the incident electron beam9. We find a structure in RNA polymerase similar to the active-site cleft of DNA polymerase I (ref. 10). In the light of functional similarities between these two enzymes, together with other evidence, this probably identifies the active-site region of RNA polymerase.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
von Hippel, P. H., Bear, D. G., Morgan, W. D. & McSwiggen, J. A. A. Rev. Biochem. 53, 389–446 (1984).
Chamberlin, M. J. Enzymes 15, 61–86 (1982).
Lewis, M. K. & Burgess, R. R. Enzymes 15, 109–153 (1982).
Allison, L. A., Moyle, M., Shales, M. & Ingles, C. J. Cell 42, 599–610 (1985).
Biggs, J., Searles, L. L. & Greenleaf, A. L. Cell 42, 611–621 (1985).
Sweetser, D., Nonet, M. & Young, R. A. Proc. natn. Acad. Sci. U.S.A. 84, 1192–1196 (1987).
Ahearn, J. M., Bartolomei, M. S., West, M. L., Cisek, L. J. & Corden, J. L. J. biol. Chem. 262, 10695–10705 (1987).
Darst, S. A., Ribi, H. O., Pierce, D. W. & Kornberg, R. D. J. molec. Biol. 203, 269–273 (1988).
Amos, L. A., Henderson, R. & Unwin, P. N. T. Prog. biophys. molec. Biol. 39, 183–231 (1982).
Ollis, D. L., Brick, P., Hamlin, R., Xuong, N. G. & Steitz, T. A. Nature 313, 762–766 (1985).
Strickland, M. S., Thompson, N. E. & Burgess, R. R. Biochemistry 27, 5755–5762 (1988).
Henderson, R., Baldwin, J. M., Downing, K. H., Lepault, J. & Zemlin, F. Ultramicroscopy 19, 147–178 (1986).
Agard, D. A. J. molec. Biol. 167, 849–852 (1983).
Tichelaar, W., Schutter, W. G., Arnberg, A. C., Van Bruggen, E. F. J. & Stender, W. Eur. J. Biochem. 135, 263–269 (1983).
Fairfield, F. R., Newport, J. W., Dolejsi, M. K. & von Hippel, P. H. J. biomolec. Struct. Dyn. 1, 715–727 (1983).
Gamper, H. B. & Hearst, J. E. Cell 29, 81–90 (1982).
Siebenlist, u., Simpson, R. B. & Gilbert, W. Cell 20, 269–281 (1980).
Kuhnke, G., Fritz, H. & Ehring, R. EMBO J. 6, 507–513 (1987).
Goodsell, D. S., Miam, I. S. & Olson, A. J. J. molec. Graphics 7, 41–47 (1989).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Darst, S., Kubalek, E. & Kornberg, R. Three-dimensional structure of Escherichia coli RNA polymerase holoenzyme determined by electron crystallography. Nature 340, 730–732 (1989). https://doi.org/10.1038/340730a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/340730a0
This article is cited by
-
Diverse and unified mechanisms of transcription initiation in bacteria
Nature Reviews Microbiology (2021)
-
Using synthetic bacterial enhancers to reveal a looping-based mechanism for quenching-like repression
Nature Communications (2016)
-
Site-specific adsorption of metallic and biological nanoparticles on nanostructured silicon surfaces
Journal of Solid State Electrochemistry (2009)
-
Eukaryotic RNA polymerase subunit RPB8 is a new relativeof the OB family
Nature Structural Biology (1998)
-
Generation of catalytic RNAs by rolling transcription of synthetic DNA nanocircles
Nature Biotechnology (1997)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.