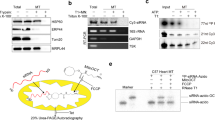
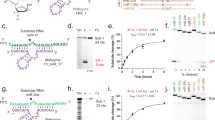
Abstract
We have been developing a unique system for the downregulation of a gene expression through cutting a specific mRNA by the long form of tRNA 3′-processing endoribonuclease (tRNase ZL) under the direction of small-guide RNA (sgRNA). However, the efficacy of this system and the involvement of tRNase ZL in the living cells were not clear. Here we show, by targeting the exogenous luciferase gene, that the efficacy of the sgRNA/tRNase ZL method can become comparable to that of the RNA interference technology and that the gene silencing is owing to tRNase ZL directed by sgRNA not owing to a simple antisense effect. We also show that tRNase ZL together with sgRNA can downregulate expression of the endogenous human genes Bcl-2 and glycogen synthase kinase-3β by degrading their mRNAs in cell culture. Furthermore, we demonstrate that a gene expression in the livers of postnatal mice can be inhibited by an only seven-nucleotide sgRNA. These data suggest that sgRNA might be utilized as therapeutic agents to treat diseases such as cancers and AIDS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Mörl M, Marchfelder A . The final cut. The importance of tRNA 3′-processing. EMBO Rep 2001; 2: 17–20.
Nashimoto M . Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res 1997; 25: 1148–1155.
Schiffer S, Rosch S, Marchfelder A . Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J 2002; 21: 2769–2777.
Takaku H, Minagawa A, Masamichi T, Nashimoto M . A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoriobonuclease. Nucleic Acids Res 2003; 31: 2272–2278.
Minagawa A, Takaku H, Takagi M, Nashimoto M . A novel endonucleolytic mechanism to generate the CCA 3′ termini of tRNA molecules in Thermotoga maritima. J Biol Chem 2004; 279: 15688–15697.
Takaku H, Minagawa A, Takagi M, Nashimoto M . The N-terminal half-domain of the long form of tRNase Z is required for the RNase 65 activity. Nucleic Acids Res 2004; 32: 4429–4438.
Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A et al. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 2001; 27: 172–180.
Zareen N, Yan H, Hopkinson A, Levinger L . Residues in the conserved His domain of fruit fly tRNase Z that function in catalysis are not involved in substrate recognition or binding. J Mol Biol 2005; 350: 189–199.
Nashimoto M . Conversion of mammalian tRNA 3′ processing endoribonuclease to four-base-recognizing RNA cutters. Nucleic Acids Res 1995; 23: 3642–3647.
Nashimoto M . Specific cleavage of target RNAs from HIV-1 with 5′ half tRNA by mammalian tRNA 3′ processing endoribonuclease. RNA 1996; 2: 2523–2524.
Nashimoto M, Geary S, Tamura M, Kasper R . RNA heptamers that directs RNA cleavage by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res 1998; 26: 2565–2571.
Nashimoto M . Anomalous RNA substrates for mammalian tRNA 3′ processing endoribonuclease. FEBS Lett 2000; 472: 179–186.
Shibata HS, Takaku H, Takagi M, Nashimoto M . The T loop structure is dispensable for substrate recognition by tRNase ZL. J Biol Chem 2005; 280: 22326–22334.
Takaku H, Minagawa A, Takagi M, Nashimoto M . A novel four-base-recognizing RNA cutter that can remove the single 3′ terminal nucleotides from RNA molecules. Nucleic Acids Res 2004; 32: e91.
Tamura M, Nashimoto C, Miyake N, Daikuhara Y, Ochi K, Nashimoto M . Intracellular mRNA cleavage by 3′ tRNase under the direction of 2′-O-methyl RNA heptamers. Nucleic Acids Res 2003; 31: 4354–4360.
HabuY, Miyano-Kurosaki N, Kitano M, Endo Y, Yukita M, Ohira S et al. Inhibition of HIV-1 gene expression by retroviral vector-mediated small-guide RNAs that direct specific RNA cleavage by tRNase ZL. Nucleic Acids Res 2005; 33: 235–243.
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T . Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001; 411: 494–498.
Hu W, Kavanagh JJ . Anticancer therapy targeting the apoptotic pathway. Lancet Oncol 2003; 4: 721–729.
Nelson WJ, Nusse R . Convergence of Wnt, beta-catenin, and cadherin pathways. Science 2004; 303: 1483–1487.
Liu F, Song Y, Liu D . Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Therapy 1999; 6: 1258–1266.
McManus MT, Sharp PA . Gene silencing in mammals by small interfering RNAs. Nat Rev Genet 2002; 3: 737–747.
Appasani K . RNA Interference Technology: from Basic Science to Drug Development. Cambridge University Press: Cambridge, 2005.
Cui J, Zhou X, LiuY, Tang Z, Romeih M . Wnt signaling in hepatocellular carcinoma: analysis of mutation and expression of beta-catenin, T-cell factor-4 and glycogen synthase kinase 3-beta genes. J Gastroenterol Hepatol 2003; 18: 280–287.
Zhang G, Budker V, Wolff JA . High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther 1999; 10: 1735–1737.
Acknowledgements
We thank Dr M Abe for creating stable transfectants. This work was supported in part by the Science Research Promotion Fund and the Academic Frontier Research Project Grant from the Promotion and Mutual Aid Corporation for Private Schools of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Gene Therapy website (http://www.nature.com/gt)
Supplementary information
Rights and permissions
About this article
Cite this article
Nakashima, A., Takaku, H., Shibata, H. et al. Gene silencing by the tRNA maturase tRNase ZL under the direction of small-guide RNA. Gene Ther 14, 78–85 (2007). https://doi.org/10.1038/sj.gt.3302841
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.gt.3302841
Keywords
This article is cited by
-
Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells
Parasitology Research (2014)
-
Identification of microRNA precursors based on random forest with network-level representation method of stem-loop structure
BMC Bioinformatics (2011)