Abstract
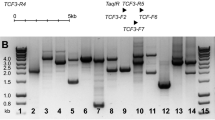
Current MRD studies in T-cell acute lymphoblastic leukemia (T-ALL) mainly use T-cell receptor gamma, delta and SIL-TAL1 gene rearrangements as MRD-PCR targets. However, low frequency or limited diversity of these markers restricts the number of evaluable patients, particularly because two markers are recommended for MRD monitoring. Hence, we developed a new strategy implementing the TCR beta (TCRB) locus for MRD quantification. The frequency and characteristics of complete and incomplete TCRB rearrangements were investigated in 53 childhood and 100 adult T-ALL patients using the BIOMED-2 multiplex PCR assay. Clonal rearrangements were identified in 92% both childhood and adult T-ALL (Vβ–Dβ–Jβ rearrangements in 80%, Dβ–Jβ rearrangements in 53%). Comparative sequence analysis of 203 TCRB recombinations revealed preferential usage of the ‘end-stage’ segment Jβ2.7 in childhood T-ALL (27%), whereas Jβ2.3 was most frequently involved in adult T-ALL (24%). In complete rearrangements, three downstream Vβ segments (19–1/20–1/21–1) were preferentially used. Subsequently, a TCRB real-time quantitative PCR assay to quantify MRD with 13 germline Jβ primer/probe combinations and allele-specific oligonucleotides was developed and applied to 60 clonal TCRB rearrangements. The assay allowed the detection of one leukemic cell within at least 104 polyclonal cells in 93% of cases and will be of high value for future MRD studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dibenedetto SP, Lo NL, Di Cataldo A, Schiliro G . Detection of minimal residual disease: methods and relationship to outcome in T-lineage acute lymphoblastic leukemia. Leukemia Lymphoma 1998; 32: 65–75.
Hoelzer D, Gokbuget N, Ottmann O, Pui CH, Relling MV, Appelbaum FR et al. Acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program) 2002; 2002: 162–192.
Cave H, van der Werff ten Bosch J, Suciu S, Guidal C, Waterkeyn C, Otten J et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer – Childhood Leukemia Cooperative Group. N Engl J Med 1998; 339: 591–598.
Coustan-Smith E, Behm FG, Sanchez J, Boyett JM, Hancock ML, Raimondi SC et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet 1998; 351: 550–554.
Coustan-Smith E, Sancho J, Hancock ML, Boyett JM, Behm FG, Raimondi SC et al. Clinical importance of minimal residual disease in childhood acute lymphoblastic leukemia. Blood 2000; 96: 2691–2696.
Dibenedetto SP, Lo NL, Mayer SP, Rovera G, Schiliro G . Detectable molecular residual disease at the beginning of maintenance therapy indicates poor outcome in children with T-cell acute lymphoblastic leukemia. Blood 1997; 90: 1226–1232.
Neale GA, Menarguez J, Kitchingman GR, Fitzgerald TJ, Koehler M, Mirro Jr J et al. Detection of minimal residual disease in T-cell acute lymphoblastic leukemia using polymerase chain reaction predicts impending relapse. Blood 1991; 78: 739–747.
van Dongen JJM, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998; 352: 1731–1738.
Willemse MJ, Seriu T, Hettinger K, d'Aniello E, Hop WC, Panzer-Grumayer ER et al. Detection of minimal residual disease identifies differences in treatment response between T-ALL and precursor B-ALL. Blood 2002; 99: 4386–4393.
Bruggemann M, Droese J, Bolz I, Luth P, Pott C, von Neuhoff N et al. Improved assessment of minimal residual disease in B cell malignancies using fluorogenic consensus probes for real-time quantitative PCR. Leukemia 2000; 14: 1419–1425.
Donovan JW, Ladetto M, Zou G, Neuberg D, Poor C, Bowers D et al. Immunoglobulin heavy-chain consensus probes for real-time PCR quantification of residual disease in acute lymphoblastic leukemia. Blood 2000; 95: 2651–2658.
Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
van der Velden VHJ, Wijkhuijs JM, Jacobs DC, van Wering ER, van Dongen JJM . T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia 2002; 16: 1372–1380.
van der Velden VHJ, Willemse MJ, van der Schoot CE, Hahlen K, van Wering ER, van Dongen JJM . Immunoglobulin kappa deleting element rearrangements in precursor-B acute lymphoblastic leukemia are stable targets for detection of minimal residual disease by real-time quantitative PCR. Leukemia 2002; 16: 928–936.
van der Velden VHJ, Hochhaus A, Cazzaniga G, Szczepanski T, Gabert J, van Dongen JJM . Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia 2003; 17: 1013–1034.
Verhagen OJ, Willemse MJ, Breunis WB, Wijkhuijs AJ, Jacobs DC, Joosten SA et al. Application of germline IGH probes in real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia. Leukemia 2000; 14: 1426–1435.
Gameiro P, Mortuza FY, Hoffbrand AV, Foroni L . Minimal residual disease monitoring in adult T-cell acute lymphoblastic leukemia: a molecular based approach using T-cell receptor g and d gene rearrangements. Haematologica 2002; 87: 1126–1134.
Nirmala K, Rajalekshmy KR, Raman SG, Shanta V, Rajkumar T . PCR-heteroduplex analysis of TCR gamma, delta and TAL-1 deletions in T-acute lymphoblastic leukemias: implications in the detection of minimal residual disease. Leuk Res 2002; 26: 335–343.
Nyvold C, Madsen HO, Ryder LP, Seyfarth J, Svejgaard A, Clausen N et al. Precise quantification of minimal residual disease at day 29 allows identification of children with acute lymphoblastic leukemia and an excellent outcome. Blood 2002; 99: 1253–1258.
van der Velden VHJ, Jacobs C, Wijkhuijs J, Comans-Bitter Willemse J, Hahle K, Kamps A et al. Minimal residual disease levels in bone marrow and peripheral blood are comparable in children with T cell acute lymphoblastic leukemia (ALL), but not in precursor-B-ALL. Leukemia 2002; 16: 1432–1436.
Szczepanski T, van der Velden VHJ, van Dongen JJM . Real-time quantitative (RQ)-PCR for the detection of minimal residual disease in childhood acute lymphoblastic leukemia. Haematologica 2002; 87: 183–191.
Breit TM, Mol EJ, Wolvers-Tettero IL, Ludwig WD, van Wering ER, van Dongen JJM . Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor alpha/beta lineage: T cell receptor delta gene deletion mechanism affects multiple genes. J Exp Med 1993; 177: 965–977.
Breit TM, Wolvers-Tettero IL, Beishuizen A, Verhoeven MA, van Wering ER, van Dongen JJM . Southern blot patterns, frequencies, and junctional diversity of T-cell receptor-delta gene rearrangements in acute lymphoblastic leukemia. Blood 1993; 82: 3063–3074.
Szczepanski T, Langerak AW, Wolvers-Tettero IL, Ossenkoppele GJ, Verhoef G, Stul M et al. Immunoglobulin and T cell receptor gene rearrangement patterns in acute lymphoblastic leukemia are less mature in adults than in children: implications for selection of PCR targets for detection of minimal residual disease. Leukemia 1998; 12: 1081–1088.
Szczepanski TP-WMJ, Langerak AW, Harts WA, Wijkhuijs AJM, van Wering ER, van Dongen JJM . Ig heavy chain gene rearrangements in T-cell acute lymphoblastic leukemia exhibit predominant DH6-19 and DH7-27 gene usage, can result in complete V–D–J rearrangements, and are rare in T-cell receptor alpha beta lineage. Blood 1999; 93: 4079–4085.
Beishuizen A, Verhoeven MA, van Wering ER, Hahlen K, Hooijkaas H, van Dongen JJM . Analysis of Ig and T-cell receptor genes in 40 childhood acute lymphoblastic leukemias at diagnosis and subsequent relapse: implications for the detection of minimal residual disease by polymerase chain reaction analysis. Blood 1994; 83: 2238–2247.
Szczepanski T, Flohr T, van der Velden, VHJ, Bartram CR, van Dongen JJM . Molecular monitoring of residual disease using antigen receptor genes in childhood acute lymphoblastic leukaemia. Best Pract Res Clin Haematol 2002; 15: 37–57.
Szczepanski T, van der Velden VHJ, Raff T, Jacobs DHJ, van Wering ER, Brüggemann M et al. Comparative analysis of T-cell receptor gene rearrangements at diagnosis and relapse of T-cell acute lymphoblastic leukemia (T-ALL) shows high stability of clonal markers for monitoring of minimal residual disease and reveals the occurrence of secondary T-ALL. Leukemia 2003; 17: 2149–2156.
van Dongen JJM, Wolvers-Tettero IL . Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta 1991; 198: 93–174.
Langerak AW, Wolvers-Tettero IL, van Dongen JJM . Detection of T cell receptor beta (TCRB) gene rearrangement patterns in T cell malignancies by Southern blot analysis. Leukemia 1999; 13: 965–974.
Assaf C, Hummel M, Dippel E, Goerdt S, Muller HH, Anagnostopoulos I et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood 2000; 96: 640–646.
Kneba M, Bolz I, Linke B, Hiddemann W . Analysis of rearranged T-cell receptor beta-chain genes by polymerase chain reaction (PCR) DNA sequencing and automated high resolution PCR fragment analysis. Blood 1995; 86: 3930–3937.
Gorski J, Yassai M, Zhu X, Kissela B, Kissella B, Keever C et al. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. Correlation with immune status. J Immunol 1994; 152: 5109–5119.
Rosenberg WM, Moss PA, Bell JI . Variation in human T cell receptor V beta and J beta repertoire: analysis using anchor polymerase chain reaction. Eur J Immunol 1992; 22: 541–549.
van Dongen JJM, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender L et al. Design and standardization of PCR primers and protocols for detection of immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations. Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Arden B, Clark SP, Kabelitz D, Mak TW . Human T-cell receptor variable gene segment families. Immunogenetics 1995; 42: 455–500.
Langerak AW, Szczepanski T, van der Burg M, Wolvers-Tettero IL, van Dongen JJM . Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997; 11: 2192–2199.
Linke B, Bolz I, Fayyazi A, von Hofen M, Pott C, Bertram J et al. Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia 1997; 11: 1055–1062.
Lefranc MP . IMGT® databases, web resources and tools for immunoglobulin and T cell receptor sequence analysis. http://imgt.cines.fr. Leukemia 2003; 17: 260–266.
Rowen L, Koop BF, Hood L . The complete 685-kilobase DNA sequence of the human beta T cell receptor locus. Science 1996; 272: 1755–1762.
Langerak AW, Wolvers-Tettero ILM, Gastel-Mol EJ, Oud MECM, van Dongen JJM . Basic helix–loop–helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood 2001; 98: 2456–2465.
Toyonaga B et al. Organization and sequences of the diversity, joining, and constant region genes of the human T-cell receptor beta chain. Proc Natl Acad Sci USA 1985; 82: 8624–8628.
Asnafi V, Beldjord K, Boulanger E, Comba B, Le Tutor P, Estienne MH et al. Analysis of TCR, pTalpha, and RAG-1 in T-acute lymphoblastic leukemias improves understanding of early human T-lymphoid lineage commitment. Blood 2003; 101: 2693–2703.
Sazawal S, Bhatia K, Gurbuxani S, Singh AL, Raina V, Khattar A et al. Pattern. pattern of immunoglobulin (Ig) and T-cell receptor (TCR) gene rearrangements in childhood acute lymphoblastic leukemia in India. Leukemia Res 2000; 24: 575–582.
Hall MA, Lanchbury JS . Healthy human T-cell receptor beta-chain repertoire. Quantitative analysis and evidence for J beta-related effects on CDR3 structure and diversity. Hum Immunol 1995; 43: 207–218.
Acknowledgements
We gratefully acknowledge Petra Chall, Petra Neumann, Frauke Hemken and Heidrun Seppelt for their technical assistance, Margot Ulrich for preparation of the figures, Sabine Hug and Regina Reutzel for providing clinical data and the German Multicenter ALL Study Group and the Dutch Childhood Leukemia Group for kindly providing the ALL samples. This work was supported by the Wilhelm Sander-Stiftung (Grant 2001.074.1), the German Compentence Network ‘Akute und Chronische Leukämien’ and by the Dutch Cancer Society/Koningin Wilhelmina Fonds (Grant SNWLK 97-1567 and Grant SNWLK 2000-2268).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Brüggemann, M., van der Velden, V., Raff, T. et al. Rearranged T-cell receptor beta genes represent powerful targets for quantification of minimal residual disease in childhood and adult T-cell acute lymphoblastic leukemia. Leukemia 18, 709–719 (2004). https://doi.org/10.1038/sj.leu.2403263
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2403263
Keywords
This article is cited by
-
Clinical features, laboratory characteristics, and outcome of ETP and TCRA/D aberrations in pediatric patients with T-acute lymphoblastic leukemia
Journal of the Egyptian National Cancer Institute (2023)
-
GvL effects in T-prolymphocytic leukemia: evidence from MRD kinetics and TCR repertoire analyses
Bone Marrow Transplantation (2017)
-
Prognostic value and clinical significance of TCR rearrangements for MRD monitoring in ALL patients
Comparative Clinical Pathology (2017)
-
Hypermethylation of p15 gene associated with an inferior poor long-term outcome in childhood acute lymphoblastic leukemia
Journal of Cancer Research and Clinical Oncology (2016)
-
Improved flow cytometric detection of minimal residual disease in childhood acute lymphoblastic leukemia
Leukemia (2013)