Abstract
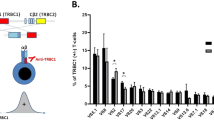
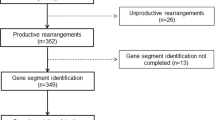
Detailed Southern blot and PCR analysis of Ig heavy (IGH), Ig kappa (IGK), T-cell receptor delta (TCRD), and TCR gamma (TCRG) genes were performed in 289 children with precursor-B-ALL in order to determine age-related Ig/TCR patterns and their implications for detection of minimal residual disease (MRD). Overall, IGH, IGK, TCRD, and TCRG gene rearrangements were detected in 98, 62, 90, and 58% of patients, respectively. The frequency of IGH and TCRD rearrangements was independent of rearrangements in one of the other three loci, whereas Ig kappa deleting element and TCRG rearrangements preferentially coincided. Southern blot analysis showed that oligoclonality of IGH, IGK, and TCRD was interrelated, that is, oligoclonality in one locus was related with a higher chance of oligoclonality in another locus. Combined Southern blot and PCR analysis revealed that Ig/TCR patterns were age related: children younger than 3 years or older than 10 years showed a higher prevalence of incomplete IGH rearrangements and a lower prevalence of IGK deletions, TCRG rearrangements, and TCRD rearrangements than children between 3 and 10 years. In addition, IGH oligoclonality was more frequent in the younger and older children. These age-related differences probably reflect ALL subsets with different cellular origin and differences in the duration of the preleukemic phase between the initial and final leukemogenetic hit. The more immature Ig/TCR gene rearrangement pattern in children younger than 3 years or older than 10 years resulted in relatively low numbers of potential MRD-PCR targets per patient, particularly if only monoclonal rearrangements were taken into account. These data provide insight into the immunobiological characteristics of Ig/TCR gene rearrangements in childhood precursor-B-ALL and form a useful basis for designing improved strategies for the identification and selection of MRD-PCR targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Szczepanski T, Orfao A, van der Velden VH, San Miguel JF, van Dongen JJ . Minimal residual disease in leukaemia patients. Lancet Oncol 2001; 2: 409–417.
Cave H, van der Werff ten Bosch J, Suciu S, Guidal C, Waterkeyn C, Otten J et al. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. European Organization for Research and Treatment of Cancer – Childhood Leukemia Cooperative Group. N Engl J Med 1998; 339: 591–598.
Coustan-Smith E, Behm FG, Sanchez J, Boyett JM, Hancock ML, Raimondi SC et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet 1998; 351: 550–554.
van Dongen JJ, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet 1998; 352: 1731–1738.
Seeger K, Kreuzer KA, Lass U, Taube T, Buchwald D, Eckert C et al. Molecular quantification of response to therapy and remission status in TEL-AML1-positive childhood ALL by real-time reverse transcription polymerase chain reaction. Cancer Res 2001; 61: 2517–2522.
Willemse MJ, Seriu T, Hettinger K, d'Aniello E, Hop WC, Panzer-Grumayer ER et al. Detection of minimal residual disease identifies differences in treatment response between T-ALL and precursor B-ALL. Blood 2002; 99: 4386–4393.
Nyvold C, Madsen HO, Ryder LP, Seyfarth J, Svejgaard A, Clausen N et al. Precise quantification of minimal residual disease at day 29 allows identification of children with acute lymphoblastic leukemia and an excellent outcome. Blood 2002; 99: 1253–1258.
Knechtli CJ, Goulden NJ, Hancock JP, Grandage VL, Harris EL, Garland RJ et al. Minimal residual disease status before allogeneic bone marrow transplantation is an important determinant of successful outcome for children and adolescents with acute lymphoblastic leukemia. Blood 1998; 92: 4072–4079.
van der Velden VH, Joosten SA, Willemse MJ, van Wering ER, Lankester AW, van Dongen JJ et al. Real-time quantitative PCR for detection of minimal residual disease before allogeneic stem cell transplantation predicts outcome in children with acute lymphoblastic leukemia. Leukemia 2001; 15: 1485–1487.
Bader P, Hancock J, Kreyenberg H, Goulden NJ, Niethammer D, Oakhill A et al. Minimal residual disease (MRD) status prior to allogeneic stem cell transplantation is a powerful predictor for post-transplant outcome in children with ALL. Leukemia 2002; 16: 1668–1672.
Schrappe M, Reiter A, Zimmermann M, Harbott J, Ludwig WD, Henze G et al. Long-term results of four consecutive trials in childhood ALL performed by the ALL-BFM study group from 1981 to 1995. Berlin–Frankfurt–Munster. Leukemia 2000; 14: 2205–2222.
Pui CH, Campana D . New definition of remission in childhood acute lymphoblastic leukemia. Leukemia 2000; 14: 783–785.
Szczepanski T, Flohr T, van der Velden VH, Bartram CR, van Dongen JJ . Molecular monitoring of residual disease using antigen receptor genes in childhood acute lymphoblastic leukaemia. Best Pract Res Clin Haematol 2002; 15: 37–57.
Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E et al. Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998; 12: 2006–2014.
Verhagen OJ, Willemse MJ, Breunis WB, Wijkhuijs AJ, Jacobs DC, Joosten SA et al. Application of germline IGH probes in real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia. Leukemia 2000; 14: 1426–1435.
Bruggemann M, Droese J, Bolz I, Luth P, Pott C, von Neuhoff N et al. Improved assessment of minimal residual disease in B cell malignancies using fluorogenic consensus probes for real-time quantitative PCR. Leukemia 2000; 14: 1419–1425.
Donovan JW, Ladetto M, Zou G, Neuberg D, Poor C, Bowers D et al. Immunoglobulin heavy-chain consensus probes for real-time PCR quantification of residual disease in acute lymphoblastic leukemia. Blood 2000; 95: 2651–2658.
van der Velden VH, Willemse MJ, van der Schoot CE, Hahlen K, van Wering ER, van Dongen JJ . Immunoglobulin kappa deleting element rearrangements in precursor-B acute lymphoblastic leukemia are stable targets for detection of minimal residual disease by real-time quantitative PCR. Leukemia 2002; 16: 928–936.
van der Velden VH, Wijkhuijs JM, Jacobs DC, van Wering ER, van Dongen JJ . T cell receptor gamma gene rearrangements as targets for detection of minimal residual disease in acute lymphoblastic leukemia by real-time quantitative PCR analysis. Leukemia 2002; 16: 1372–1380.
Szczepanski T, van der Velden VH, van Dongen JJ . real-time quantitative (RQ)-PCR for the detection of minimal residual disease in childhood acute lymphoblastic leukemia. Haematologica 2002; 87: 183–191.
van Dongen JJ, Wolvers-Tettero IL . Analysis of immunoglobulin and T cell receptor genes. Part I: basic and technical aspects. Clin Chim Acta 1991; 198: 1–91.
de Haas V, Verhagen OJ, von dem Borne AE, Kroes W, van den Berg H, van der Schoot CE . Quantification of minimal residual disease in children with oligoclonal B-precursor acute lymphoblastic leukemia indicates that the clones that grow out during relapse already have the slowest rate of reduction during induction therapy. Leukemia 2001; 15: 134–140.
Moreira I, Papaioannou M, Mortuza FY, Gameiro P, Palmisano GL, Harrison CJ et al. Heterogeneity of VH–JH gene rearrangement patterns: an insight into the biology of B cell precursor ALL. Leukemia 2001; 15: 1527–1536.
Szczepanski T, Willemse MJ, Brinkhof B, van Wering ER, van der Burg M, van Dongen JJ . Comparative analysis of Ig and TCR gene rearrangements at diagnosis and at relapse of childhood precursor-B-ALL provides improved strategies for selection of stable PCR targets for monitoring of minimal residual disease. Blood 2002; 99: 2315–2323.
Steward CG, Goulden NJ, Katz F, Baines D, Martin PG, Langlands K et al. A polymerase chain reaction study of the stability of Ig heavy-chain and T-cell receptor delta gene rearrangements between presentation and relapse of childhood B-lineage acute lymphoblastic leukemia. Blood 1994; 83: 1355–1362.
Kitchingman GR . Immunoglobulin heavy chain gene VH–D junctional diversity at diagnosis in patients with acute lymphoblastic leukemia. Blood 1993; 81: 775–782.
Bird J, Galili N, Link M, Stites D, Sklar J . Continuing rearrangement but absence of somatic hypermutation in immunoglobulin genes of human B cell precursor leukemia. J Exp Med 1988; 168: 229–245.
Beishuizen A, Verhoeven MA, van Wering ER, Hahlen K, Hooijkaas H, van Dongen JJ . Analysis of Ig and T-cell receptor genes in 40 childhood acute lymphoblastic leukemias at diagnosis and subsequent relapse: implications for the detection of minimal residual disease by polymerase chain reaction analysis. Blood 1994; 83: 2238–2247.
Brumpt C, Delabesse E, Beldjord K, Davi F, Cayuela JM, Millien C et al. The incidence of clonal T-cell receptor rearrangements in B-cell precursor acute lymphoblastic leukemia varies with age and genotype. Blood 2000; 96: 2254–2261.
Szczepanski T, Langerak AW, Wolvers-Tettero IL, Ossenkoppele GJ, Verhoef G, Stul M et al. Immunoglobulin and T cell receptor gene rearrangement patterns in acute lymphoblastic leukemia are less mature in adults than in children: implications for selection of PCR targets for detection of minimal residual disease. Leukemia 1998; 12: 1081–1088.
Nuss R, Kitchingman G, Cross A, Zipf TF, Antoun GR, Bernstein I et al. T cell receptor gene rearrangements in B-precursor acute lymphoblastic leukemia correlate with age and the stage of B cell differentiation. Leukemia 1988; 2: 722–727.
Peham M, Panzer S, Fasching K, Haas OA, Fischer S, Marschalek R et al. Low frequency of clonotypic Ig and T-cell receptor gene rearrangements in t(4;11) infant acute lymphoblastic leukaemia and its implication for the detection of minimal residual disease. Br J Haematol 2002; 117: 315–321.
Szczepanski T, Willemse MJ, van Wering ER, van Weerden JF, Kamps WA, van Dongen JJ . Precursor-B-ALL with D(H)–J(H) gene rearrangements have an immature immunogenotype with a high frequency of oligoclonality and hyperdiploidy of chromosome 14. Leukemia 2001; 15: 1415–1423.
Breit TM, Mol EJ, Wolvers-Tettero IL, Ludwig WD, van Wering ER, van Dongen JJ . Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor alpha/beta lineage: T cell receptor delta gene deletion mechanism affects multiple genes. J Exp Med 1993; 177: 965–977.
Felix CA, Reaman GH, Korsmeyer SJ, Hollis GF, Dinndorf PA, Wright JJ et al. Immunoglobulin and T cell receptor gene configuration in acute lymphoblastic leukemia of infancy. Blood 1987; 70: 536–541.
Verhagen OJ, Wijkhuijs AJ, van der Sluijs-Gelling AJ, Szczepanski T, van der Linden-Schrever BE, Pongers-Willemse MJ et al. Suitable DNA isolation method for the detection of minimal residual disease by PCR techniques. Leukemia 1999; 13: 1298–1299.
Beishuizen A, Verhoeven MA, Mol EJ, van Dongen JJ . Detection of immunoglobulin kappa light-chain gene rearrangement patterns by Southern blot analysis. Leukemia 1994; 8: 2228–2236.
Beishuizen A, Verhoeven MA, Mol EJ, Breit TM, Wolvers-Tettero IL, van Dongen JJ . Detection of immunoglobulin heavy-chain gene rearrangements by Southern blot analysis: recommendations for optimal results. Leukemia 1993; 7: 2045–2053.
Breit TM, Wolvers-Tettero IL, Beishuizen A, Verhoeven MA, van Wering ER, van Dongen JJ . Southern blot patterns, frequencies, and junctional diversity of T-cell receptor-delta gene rearrangements in acute lymphoblastic leukemia. Blood 1993; 82: 3063–3074.
Moreau EJ, Langerak AW, van Gastel-Mol EJ, Wolvers-Tettero IL, Zhan M, Zhou Q et al. Easy detection of all T cell receptor gamma (TCRG) gene rearrangements by Southern blot analysis: recommendations for optimal results. Leukemia 1999; 13: 1620–1626.
Szczepanski T, Pongers-Willemse MJ, Langerak AW, van Dongen JJ . Unusual immunoglobulin and T-cell receptor gene rearrangement patterns in acute lymphoblastic leukemias. Curr Top Microbiol Immunol 1999; 246: 205–213; discussion 214–205.
Pongers-Willemse MJ, Seriu T, Stolz F, d'Aniello E, Gameiro P, Pisa P et al. Primers and protocols for standardized detection of minimal residual disease in acute lymphoblastic leukemia using immunoglobulin and T cell receptor gene rearrangements and TAL1 deletions as PCR targets: report of the BIOMED-1 CONCERTED ACTION: investigation of minimal residual disease in acute leukemia. Leukemia 1999; 13: 110–118.
Szczepanski T, Pongers-Willemse MJ, Langerak AW, Harts WA, Wijkhuijs AJ, van Wering ER et al. Ig heavy chain gene rearrangements in T-cell acute lymphoblastic leukemia exhibit predominant DH6-19 and DH7-27 gene usage, can result in complete V–D–J rearrangements, and are rare in T-cell receptor alpha beta lineage. Blood 1999; 93: 4079–4085.
Langerak AW, Szczepanski T, van der Burg M, Wolvers-Tettero IL, van Dongen JJ . Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997; 11: 2192–2199.
Boeckx N, Willemse MJ, Szczepanski T, van der Velden VH, Langerak AW, Vandekerckhove P et al. Fusion gene transcripts and Ig/TCR gene rearrangements are complementary but infrequent targets for PCR-based detection of minimal residual disease in acute myeloid leukemia. Leukemia 2002; 16: 368–375.
van der Burg M, Barendregt BH, Szczepanski T, van Wering ER, Langerak AW, van Dongen JJ . Immunoglobulin light chain gene rearrangements display hierarchy in absence of selection for functionality in precursor-B-ALL. Leukemia 2002; 16: 1448–1453.
Szczepanski T, Beishuizen A, Pongers-Willemse MJ, Hahlen K, Van Wering ER, Wijkhuijs AJ et al. Cross-lineage T cell receptor gene rearrangements occur in more than ninety percent of childhood precursor-B acute lymphoblastic leukemias: alternative PCR targets for detection of minimal residual disease. Leukemia 1999; 13: 196–205.
Greaves MF, Chan LC, Furley AJ, Watt SM, Molgaard HV . Lineage promiscuity in hemopoietic differentiation and leukemia. Blood 1986; 67: 1–11.
van Dongen JJ, Wolvers-Tettero IL . Analysis of immunoglobulin and T cell receptor genes. Part II: possibilities and limitations in the diagnosis and management of lymphoproliferative diseases and related disorders. Clin Chim Acta 1991; 198: 93–174.
Sorensen PH, Chen CS, Smith FO, Arthur DC, Domer PH, Bernstein ID et al. Molecular rearrangements of the MLL gene are present in most cases of infant acute myeloid leukemia and are strongly correlated with monocytic or myelomonocytic phenotypes. J Clin Invest 1994; 93: 429–437.
van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia 1999; 13: 1901–1928.
Fasching K, Panzer S, Haas OA, Borkhardt A, Marschalek R, Griesinger F et al. Presence of N regions in the clonotypic DJ rearrangements of the immunoglobulin heavy-chain genes indicates an exquisitely short latency in t(4;11)-positive infant acute lymphoblastic leukemia. Blood 2001; 98: 2272–2274.
Biondi A, Cimino G, Pieters R, Pui CH . Biological and therapeutic aspects of infant leukemia. Blood 2000; 96: 24–33.
Gill Super HJ, Rothberg PG, Kobayashi H, Freeman AI, Diaz MO, Rowley JD . Clonal, nonconstitutional rearrangements of the MLL gene in infant twins with acute lymphoblastic leukemia: in utero chromosome rearrangement of 11q23. Blood 1994; 83: 641–644.
Chessells JM, Hall E, Prentice HG, Durrant J, Bailey CC, Richards SM . The impact of age on outcome in lymphoblastic leukaemia; MRC UKALL X and XA compared: a report from the MRC Paediatric and Adult Working Parties. Leukemia 1998; 12: 463–473.
Donadieu J, Auclerc MF, Baruchel A, Leblanc T, Landman-Parker J, Perel Y et al. Critical study of prognostic factors in childhood acute lymphoblastic leukaemia: differences in outcome are poorly explained by the most significant prognostic variables. Fralle group. French Acute Lymphoblastic Leukaemia study group. Br J Haematol 1998; 102: 729–739.
Gaynon PS, Crotty ML, Sather HN, Bostrom BC, Nachman JB, Steinherz PG et al. Expression of BCR-ABL, E2A-PBX1, and MLL-AF4 fusion transcripts in newly diagnosed children with acute lymphoblastic leukemia: a Children's Cancer Group initiative. Leukamia Lymphoma 1997; 26: 57–65.
Akashi K, Taniguchi S, Nagafuji K, Harada M, Shibuya T, Hayashi S et al. B-lymphoid/myeloid stem cell origin in Ph-positive acute leukemia with myeloid markers. Leukamia Res 1993; 17: 549–555.
Lo Coco F, Basso G, di Celle PF, Tassinari A, Pasqualetti D, De Cuia MR et al. Molecular characterization of Ph'+hybrid acute leukemia. Leukemia Res 1989; 13: 1061–1067.
Raimondi SC, Pui CH, Hancock ML, Behm FG, Filatov L, Rivera GK . Heterogeneity of hyperdiploid (51–67) childhood acute lymphoblastic leukemia. Leukemia 1996; 10: 213–224.
Greaves M . Molecular genetics, natural history and the demise of childhood leukaemia. Eur J Cancer 1999; 35: 1941–1953.
Panzer-Grumayer ER, Fasching K, Panzer S, Hettinger K, Schmitt K, Stockler-Ipsiroglu S et al. Nondisjunction of chromosomes leading to hyperdiploid childhood B-cell precursor acute lymphoblastic leukemia is an early event during leukemogenesis. Blood 2002; 100: 347–349.
Taub JW, Konrad MA, Ge Y, Naber JM, Scott JS, Matherly LH et al. High frequency of leukemic clones in newborn screening blood samples of children with B-precursor acute lymphoblastic leukemia. Blood 2002; 99: 2992–2996.
Williams DL, Look AT, Melvin SL, Roberson PK, Dahl G, Flake T et al. New chromosomal translocations correlate with specific immunophenotypes of childhood acute lymphoblastic leukemia. Cell 1984; 36: 101–109.
Raimondi SC, Behm FG, Roberson PK, Williams DL, Pui CH, Crist WM et al. Cytogenetics of pre-B-cell acute lymphoblastic leukemia with emphasis on prognostic implications of the t(1;19). J Clin Oncol 1990; 8: 1380–1388.
Langerak AW, Wolvers-Tettero IL, van Gastel-Mol EJ, Oud ME, van Dongen JJ . Basic helix–loop–helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood 2001; 98: 2456–2465.
Wiemels JL, Leonard BC, Wang Y, Segal MR, Hunger SP, Smith MT et al. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA 2002; 99: 15101–15106.
Acknowledgements
We acknowledge Marieke Comans-Bitter for preparation of the figures and Maaike de Bie for technical assistance. We gratefully acknowledge Dr Anton W Langerak, Dr Mirjam van der Burg, Dr Mieke Jansen, and Professor Dr Elisabeth Macintyre for helpful discussions and for critically reviewing the manuscript. We thank the Dutch Childhood Oncology Group for kindly providing precursor-B-ALL cell samples.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
van der Velden, V., Szczepanski, T., Wijkhuijs, J. et al. Age-related patterns of immunoglobulin and T-cell receptor gene rearrangements in precursor-B-ALL: implications for detection of minimal residual disease. Leukemia 17, 1834–1844 (2003). https://doi.org/10.1038/sj.leu.2403038
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2403038
Keywords
This article is cited by
-
Minimal residual disease detection by next-generation sequencing of different immunoglobulin gene rearrangements in pediatric B-ALL
Nature Communications (2023)
-
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines
BMC Cancer (2018)
-
Prognostic value and clinical significance of TCR rearrangements for MRD monitoring in ALL patients
Comparative Clinical Pathology (2017)
-
Minimal residual disease in acute lymphoblastic leukemia: optimal methods and clinical relevance, pitfalls and recent approaches
Medical Oncology (2014)
-
Extremely low-frequency magnetic fields and survival from childhood acute lymphoblastic leukemia: an international follow-up study
Blood Cancer Journal (2012)