Abstract
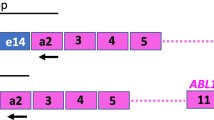
The inv(16)(p13q22) chromosomal rearrangement associated with FAB M4Eo acute myeloid leukemia (AML) subtype is characterized by the presence of the CBFbeta/MYH11 fusion transcript that can be used to detect minimal residual disease (MRD). However, qualitative RT-PCR studies of MRD have so far produced conflicting results and seem of limited prognostic value. We have evaluated retrospectively MRD in a large series of CBFbeta/MYH11-positive patients employing both qualitative and quantitative (real-time PCR) approaches. 186 bone marrow samples from 36 patients were examined with a median follow-up of 27.5 months; 15 patients relapsed during follow-up. In qualitative studies, carried out by ‘nested’ RT-PCR assay, all patients in complete remission (CR) immediately after induction/consolidation therapy were found to be PCR positive. However, follow-up samples at later time points were persistently negative (except one case) in patients remaining in continuous CR (CCR) for more than 12 months. 16 patients were evaluated by quantitative real-time PCR assay: CBFbeta/MYH11 transcript copy number was normalized for expression of the housekeeping gene ABL, expressed as fusion gene copy number per 104 copies of ABL. A 2–3 log decline in leukemic transcript copy number was observed after induction/consolidation therapy. After achieving CR, the mean copy number was significantly higher in patients destined to relapse compared to patients remaining in CCR (151 vs 9, P < 0.0001 by Mann–Whitney test). Moreover, in CCR patients, the copy number dropped below the detection threshold after the treatment protocol was completed and remained undetectable in subsequent MRD analysis in accordance with results obtained by qualitative RT-PCR. On the contrary, in the seven patients who relapsed, the copy number in CR never declined below the detection threshold; thus a cut-off value discriminating these two groups of patients could be established. The findings of our study, if confirmed, might confer an important predictive value to quantitative real-time PCR determinations of MRD in patients with inv(16) leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bitter MA, Le Beau MM, Rowley JD, Larson RA, Golomb HM, Vardiman JW . Association between morphology, karyotype, and clinical features in myeloid leukemias Hum Pathol 1987 18: 211–225
Mrozek K, Heinonen K, de la Chapelle A, Bloomfield CD . Clinical significance of cytogenetics in acute myeloid leukemia Semin Oncol 1997 24: 17
Rowe D, Cotterill SJ, Ross FM, Bunyan DJ, Vickers SJ, Bryon J, McMullan DJ, Griffits MJ, Reilly JT, Vandenberghe EA, Wilson G, Watmore AE, Bown NP . Cytogenetically cryptic AML1-ETO and CBFbeta-MYH11 gene rearrangement: incidence in 412 cases of acute myeloid leukaemia Br J Haematol 2000 111: 1051–1056
Downing JR . AML1/CBFbeta transcription complex: its role in normal hematopoiesis and leukemia Leukemia 2001 15: 664–665
Liu P, Hajra A, Wijmenga C, Collins FS . Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia Blood 1995 85: 2289–2302
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A on behalf of the Medical Research Council Adult and Children's Leukaemia Working Parties. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1612 patients entered into the MRC AML 10 trial Blood 1998 92: 2322–2333
Lowenberg B . Postremission treatment of acute myelogenous leukemia N Eng J Med 1995 332: 260–262
Claxton DF, Liu P, Hsu HB, Marlton P, Hester J, Collins F, Deisseroth AB, Rowley JD, Siciliano MJ . Detection of fusion transcripts generated by the inversion 16 chromosome in acute myelogenous leukemia Blood 1994 83: 1750
Hébert J, Cayuela JM, Daniel MT, Berger R, Sigaux F . Detection of minimal residual disease in acute myelomonocytic leukemia with abnormal marrow eosinophils by nested polymerase chain reaction with allele specific amplification Blood 1994 84: 2291–2296
Poirel H, Radford-Weiss I, Rack K, Troussard X, Veil A, Valensi F, Picard F, Guesnu M, Leboeuf D, Melle J . Detection of the chromosome 16 CBF-MYH11 fusion transcript in myelomonocytic leukemias Blood 1995 85: 1313–1322
Guerrasio A, Rosso C, Martinelli G, Lo Coco F, Pampinella M, Santoro A, Lanza A, Allione B, Resegotti L, Saglio G . Polyclonal hematopoiesis associated with long-term persistence of AML1-ETO transcript in patients with FAB M2 acute myeloid leukemia in continuous clinical remission Br J Haematol 1995 90: 364–368
Elmaagacli AH, Beelen DW, Stockowa J, Trensky S, Kroll M, Schaefer UW, Stein C, Opalka B . Detection of AML1-ETO fusion transcripts in patients with t(8;21) acute myeloid leukemia after allogeneic bone marrow transplantation or peripheral blood progenitor cell transplantation Blood 1997 90: 3230–3231
Jurlander J, Caligiuri MA, Ruutu T, Baer MR, Strout MP, Oberkircher AR, Hoffmann L, Ball ED, Frei-Lahr DA, Christiansen NP, Block AM, Knuutila S, Herzig GP, Bloomfield CD . Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia Blood 1996 88: 2183
Morschhauser F, Cayuela JM, Martini S, Baruchel A, Rousselot P, Sociè G, Berthou P, Jouet JP, Straetmans N, Sigaux F, Fenaux P, Preudhomme C . Evaluation of minimal residual disease using reverse-transcription polymerase chain reaction in t(8;21) acute myeloid leukemia: a multicenter study of 51 patients J Clin Oncol 2000 18: 778–794
Tobal K, Johnson PR, Saunders MJ, Harrison CJ, Liu Yin JA . Detection of CBFB/MYH11 transcripts in patients with inversion and other abnormalities of chromosome 16 at presentation and remission Br J Haematol 1995 91: 104
Costello R, Sainty D, Blaise D, Gastaut JA, Gabert J . Prognosis value of residual disease monitoring by polymerase chain reaction in patients with CBF/MYH11-positive acute myeloblastic leukemia Blood 1997 89: 2222 (letter)
Marcucci G, Caligiuri MA, Bloomfield CD . Defining the ‘absence’ of CBFbeta/MYH11 fusion transcript in patients with acute myeloid leukemia and inversion of chromosome 16 to predict long term complete remission: a call for definition Blood 1997 90: 5022–5024
Evans PA, Short MA, Jack AS, Norfolk DR, Child JA, Shiach CR, Davies F, Tobal K, Liu Yin JA, Morgan GJ . Detection and quantitation of the transcripts associated with the inv(16) in presentation and follow-up samples from patients with AML Leukemia 1997 11: 364
Laczika K, Novak M, Hilgarth B, Mitterbauer M, Mitterbauer G, Scheidel-Petrovic A, Scholten C, Thalammer-Sherrer R, Brugger S, Keil F, Schwarzinger I, Haas OA, Lechner K, Jaeger U . Competitive CBFbeta/MYH11 reverse transcriptase polymerase chain reaction for quantitative assessment of minimal residual disease during post remission therapy in acute myeloid leukemia with inversion 16: a pilot study J Clin Oncol 1998 16: 1519–1525
Krauter J, Hoellge W, Wattjies MP, Nagel S, Heidenreich O, Bunjes D, Ganser A, Heil G . Detection and quantitation of CBFB/MYH11 fusion transcript in patients with inv(16) positive acute myeloblastic leukemia by real-time RT-PCR Genes Chromos Cancer 2001 30: 342–348
Marcucci G, Caligiuri MA, Dohner H, Archer KJ, Schlenk RF, Dohner K, Maghraby EA, Bloomfield CD . Quantification of CBFbeta/MYH11 fusion trancript by real-time RT-PCR in patients with inv(16) acute myeloid leukemia Leukemia 2001 15: 1072–1080
Van Dongen JJM, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, Gottardi E, Rambaldi A, Dotti G, Griesinger F, Parreira A, Gameiro P, Gonzàlez Diàz M, Malec M, Langerak AW, San Miguel JF, Biondi A . Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease Leukemia 1999 13: 1901–1928
Gabert J, Beillard E, Bi W, Pallisgaard N, Gottardi E, Cazzaniga G, Barbany G, Cavè H, Cayuela JM, Grimwade D, Aerts J, Van Der Velden V, Pane F, Saglio G, Van Dongen JJM . On behalf of the European SANCO Concerted Action: European standardization and quality control program of real-time quantitative RT-PCR analysis of fusion gene transcripts for minimal residual disease detection in leukemia patients Blood 2000 96: 1343 (Abstr.)
Martin G, Barragan E, Bolufer P, Chillon C, Garcia-Sanz R, Gomez T, Brunet S, Gonzalez M, Sanz MA . Relevance of presenting white blood cell count and kinetics of molecular remission in the prognosis of acute myeloid leukemia with CBFbeta/MYH11 rearrangement Haematologica 2000 85: 699–703
Beillard E, van der Velden V, Bi W, Pallisgaard N, De Micheli D, Cazzaniga G, Barbany G, Cayuela JM, Pane F, Cavé H, Grimwade D, Aerts JLE, Thirion X, Pradel V, González M, Viehmann S, Malec M, Saglio G, van Dongen JJM, Gabert J . Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction (RQ-PCR) of fusion gene transcripts for minimal residual disease detection in leukemia – A Europe Against Cancer Program Leukemia (submitted)
Pallisgaard N, Hokland P, Bi W, Dee R, van der Schoot E, Delabesse E, Macintyre E, van der Velden V, van Dongen JJM, Gottardi E, Saglio G, Watzinger F, Leon T, Beillard E, Gabert J . Control genes for real-time quantitative RT-PCR in leukemia. A report from SANCO Concerted Action within the Europe Against Cancer Program Leukemia (submitted)
Acknowledgements
This work was supported by grants from CNR (Progetto Finalizzato Biotecnologie), MURST-COFIN 2000, AIL (Associazione Italiana contro le Leucemie) and AIRC (Associazione Italiana per la Ricerca sul Cancro). DC is supported by fellowship of the Associazione Italiana Amici di Josè Carreras. We thank the European SANCO Group for useful collaboration in implementing PCR technology. We are indebted to Dr E Aruta for statistical assistance.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Guerrasio, A., Pilatrino, C., De Micheli, D. et al. Assessment of minimal residual disease (MRD) in CBFbeta/MYH11-positive acute myeloid leukemias by qualitative and quantitative RT-PCR amplification of fusion transcripts. Leukemia 16, 1176–1181 (2002). https://doi.org/10.1038/sj.leu.2402478
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.leu.2402478