Abstract
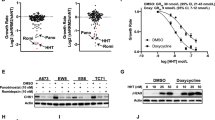
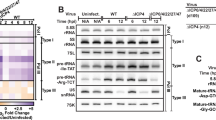
DDX3 is a human RNA helicase with plethoric functions. Our previous studies have indicated that DDX3 is a transcriptional regulator and functions as a tumor suppressor. In this study, we use a bicistronic reporter to demonstrate that DDX3 specifically represses cap-dependent translation but enhances hepatitis C virus internal ribosome entry site-mediated translation in vivo in a helicase activity-independent manner. To elucidate how DDX3 modulates translation, we identified translation initiation factor eukaryotic initiation factor 4E (eIF4E) as a DDX3-binding partner. Interestingly, DDX3 utilizes a consensus eIF4E-binding sequence YIPPHLR to interact with the functionally important dorsal surface of eIF4E in a similar manner to other eIF4E-binding proteins. Furthermore, cap affinity chromatography analysis suggests that DDX3 traps eIF4E in a translationally inactive complex by blocking interaction with eIF4G. Point mutations within the consensus eIF4E-binding motif in DDX3 impair its ability to bind eIF4E and result in a loss of DDX3's regulatory effects on translation. All these features together indicate that DDX3 is a new member of the eIF4E inhibitory proteins involved in translation initiation regulation. Most importantly, this DDX3-mediated translation regulation also confers the tumor suppressor function on DDX3. Altogether, this study demonstrates regulatory roles and action mechanisms for DDX3 in translation, cell growth and likely viral replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Andrei MA, Ingelfinger D, Heintzmann R, Achsel T, Rivera-Pomar R, Luhrmann R . (2005). A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11: 717–727.
Chang PC, Chi CW, Chau GY, Li FY, Tsai YH, Wu JC et al. (2006). DDX3, a DEAD box RNA helicase, is deregulated in hepatitis virus-associated hepatocellular carcinoma and is involved in cell growth control. Oncogene 25: 1991–2003.
Chao CH, Chen CM, Cheng PL, Shih JW, Tsou AP, Wu Lee YH . (2006). DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res 66: 6579–6588.
Chuang RY, Weaver PL, Liu Z, Chang TH . (1997). Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275: 1468–1471.
De Benedetti A, Graff JR . (2004). eIF-4E expression and its role in malignancies and metastases. Oncogene 23: 3189–3199.
de la Cruz J, Iost I, Kressler D, Linder P . (1997). The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94: 5201–5206.
Ferraiuolo MA, Basak S, Dostie J, Murray EL, Schoenberg DR, Sonenberg N . (2005). A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol 170: 913–924.
Gebauer F, Hentze MW . (2004). Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 5: 827–835.
Gingras AC, Raught B, Sonenberg N . (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963.
Hellen CU, Sarnow P . (2001). Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612.
Horton LE, Bushell M, Barth-Baus D, Tilleray VJ, Clemens MJ, Hensold JO . (2002). p53 activation results in rapid dephosphorylation of the eIF4E-binding protein 4E-BP1, inhibition of ribosomal protein S6 kinase and inhibition of translation initiation. Oncogene 21: 5325–5334.
Hwang LH, Hsieh CL, Yen A, Chung YL, Chen DS . (1998). Involvement of the 5′ proximal coding sequences of hepatitis C virus with internal initiation of viral translation. Biochem Biophys Res Commun 252: 455–460.
Kanai Y, Dohmae N, Hirokawa N . (2004). Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43: 513–525.
Kao CF, Chen SY, Chen JY, Wu Lee YH . (2004). Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene 23: 2472–2483.
Linder P . (2003). Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol Cell 95: 157–167.
Lynch M, Fitzgerald C, Johnston KA, Wang S, Schmidt EV . (2004). Activated eIF4E-binding protein slows G1 progression and blocks transformation by c-myc without inhibiting cell growth. J Biol Chem 279: 3327–3339.
Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N . (2004). eIF4E--from translation to transformation. Oncogene 23: 3172–3179.
Mamiya N, Worman HJ . (1999). Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274: 15751–15756.
Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK . (1999). Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3: 707–716.
Niessing D, Blanke S, Jackle H . (2002). Bicoid associates with the 5′-cap-bound complex of caudal mRNA and represses translation. Genes Dev 16: 2576–2582.
Ohlmann T, Rau M, Pain VM, Morley SJ . (1996). The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J 15: 1371–1382.
Owsianka AM, Patel AH . (1999). Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology 257: 330–340.
Park SH, Lee SG, Kim Y, Song K . (1998). Assignment of a human putative RNA helicase gene, DDX3, to human X chromosome bands p11.3 → p11.23. Cytogenet Cell Genet 81: 178–179.
Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence Jr JC et al. (1994). Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371: 762–767.
Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N . (1998). 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 273: 14002–14007.
Richter JD, Sonenberg N . (2005). Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480.
Rocak S, Linder P . (2004). DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241.
Rousseau D, Gingras AC, Pause A, Sonenberg N . (1996). The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13: 2415–2420.
Sachs AB, Varani G . (2000). Eukaryotic translation initiation: there are (at least) two sides to every story. Nat Struct Biol 7: 356–361.
Strudwick S, Borden KL . (2002). The emerging roles of translation factor eIF4E in the nucleus. Differentiation 70: 10–22.
Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N . (2005). Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol Cell Biol 25: 10556–10565.
Tanner NK, Linder P . (2001). DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell 8: 251–262.
von der Haar T, Gross JD, Wagner G, McCarthy JE . (2004). The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 11: 503–511.
Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S et al. (2003). The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23: 26–37.
Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT . (2004). Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392.
You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH et al. (1999). Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol 73: 2841–2853.
Zhou Z, Licklider LJ, Gygi SP, Reed R . (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185.
Acknowledgements
We are grateful to Dr A Lin and IJ Chen for assistance in polysome profile analysis. We thank Drs MR Chen and LH Hwang for generously providing experimental materials, and Dr R Kirby for critical reading and comments on this paper. This work was supported by the following grants to Dr Y-H Wu Lee: NSC-92-2320-B-010-014, NSC-93-2320-B-010-049, NSC-94-2320-B-010-053 and NSC-95-2320-B-010-012 from the National Science Council as well as NHRI-EX94-9002BL, NHRI-EX95-9501BI and NHRI-EX96-9501BI from the National Health Research Institute and the ‘Aim for the Top University Plan’ grant from the Ministry of Education of the Republic of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Supplementary information
Rights and permissions
About this article
Cite this article
Shih, JW., Tsai, TY., Chao, CH. et al. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27, 700–714 (2008). https://doi.org/10.1038/sj.onc.1210687
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210687
Keywords
This article is cited by
-
Molecular docking, synthesis, and biological evaluation of 7-azaindole-derivative (7AID) as novel anti-cancer agent and potent DDX3 inhibitor:—an in silico and in vitro approach
Medical Oncology (2022)
-
DDX3X: structure, physiologic functions and cancer
Molecular Cancer (2021)
-
The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis
International Journal of Clinical Oncology (2021)
-
Control of the eIF4E activity: structural insights and pharmacological implications
Cellular and Molecular Life Sciences (2021)
-
Targeting mitochondrial translation by inhibiting DDX3: a novel radiosensitization strategy for cancer treatment
Oncogene (2018)