Abstract
Tay-Sachs disease is a lipidosis due to the deficiency of the lysosomal hexosaminidase A. In order to understand the molecular mechanisms of this enzyme deficiency we studied 42 patients of different ethnic origins diagnosed in Europe. The strategy used consists in HEXA cDNA amplification followed by allele-specific oligonucleotide analysis for the frequent mutations, and by chemical cleavage mismatch and denaturing gradient gel electrophoresis for the detection of new mutations. 90% of alleles were clarified in this way, showing a high heterogeneity of HEXA lesions in Tay-Sachs disease. 28 different mutations were found, 20 being identified for the first time in this group of patients.
Similar content being viewed by others
Identification of gene lesions responsible for a genetic disease can have several aims: to understand the gene function, to reveal the pathogenic mechanisms involved, to correlate a mutation with a specific phenotype or to clarify the molecular epidemiology of a particular disease in a particular population. To reach these objectives we undertook a study of the distribution of mutations responsible for Tay-Sachs disease in the European population.
Previous investigations have shown that different mutations of the HEXA gene can produce defects in the a subunit of β-hexosaminidase A, which is responsible for Tay-Sachs disease. The loss of this activity results in a lysosomal accumulation of undegraded GM2 ganglioside, particularly in neurons. Clinically, the disorder displays varying degrees of severity [1] related to the level of residual β-hexosaminidase A activity toward its natural substrate [2, 3].
The effort to identify alleles in Tay-Sachs disease originally focused on the Ashkenazi Jewish and French-Canadian populations showing a high incidence of the disease. In Ashkenazi Jews, two major mutations responsible for the infantile form have been demonstrated: a G to C transversion at the 5′ splice site of intron 12 [4–6], and a 4-bp insertion in exon 11 [7], also found among non-Jewish patients [8]. The adult form of Tay-Sachs disease is characterized by a point mutation at the 3′ end of exon 7 that results in the substitution of Ser for Gly at codon 269 [9, 10]. Among French-Canadians, a 7.6-kb deletion at the 5′ end of the gene [11] accounts for about 80% of mutant chromosomes examined in this ethnic group [12].
Other mutations have been reported in the literature in families from a diversity of ethnic backgrounds, with most of these mutations being described in just one family [13]. Such a heterogeneity of mutations emphasizes the need to test screening strategies to detect simultaneously known and unknown mutations.
We give here a survey of conclusions resulting from the study of 42 Tay-Sachs patients of different ethnic origins diagnosed in our different European laboratories. The strategy used allowed us to clarify 90% of alleles investigated and to establish genotype-phenotype and epidemiological correlations.
Material and Methods
Strategy
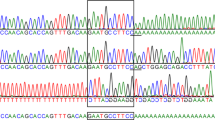
The strategy used in this study is based upon the analysis of mRNA which is the most compact version of a gene and has the advantage of allowing the detection of splice mutations in the form of aberrant or truncated transcripts (exon skipping). After conversion into single-strand cDNA, the HEXA coding sequence was PCR-amplified in two overlapping fragments [see ref. 14 for primer sequence]. We first screened all the patients by specific oligonucleotides for the most frequent known mutations [allele-specific oligonucleotide (ASO) analysis; see table 1]. Then we chose chemical mismatch cleavage (CMC) analysis [15] to screen the entire coding region for mutations in all 42 patients, and denaturing gradient gel electrophoresis (DGGE) analysis of a 5′ cDNA segment spanning the active site for 19 of these patients. The CMC method detects all classes of mutations [15] and permits localization of the mutation limiting DNA sequencing to only the region of mismatch. DGGE is very efficient for detecting all types of mutations but does not precisely locate the sequence alteration and is restricted to the analysis of small DNA segments up to 400 bp because of the limited size of melting domains [see fig. 1]. To determine whether the patients were homozygotes or heterozygotes for each mutation the corresponding regions of genomic DNA were sequenced. The deletion characteristic of the French-Canadian infantile form was screened by PCR with 3 oligoprimers as described elsewhere [16].
Results of CMC and DGGE of human HEXA cDNA on patients with Tay-Sachs disease. a DGGE analysis of the fragment spanning exon 1 to exon 7 with a GC clamp attached to the 5′ end. The results of one control (N) and 4 subjects with extra bands are shown. When samples have a single band, we mix normal and patient DNA in a ratio of 1:1 to form heteroduplexes (lane KM + N). b Autoradiograph with cleavage products. In all lanes the full-length probe of 461 bp is seen. Lanes 3 and 11 are from patients DA and NA, respectively, after hydroxylamine modification. Lane 1 = probe alone; lane 2 = normal cDNA control (no cleavage). Other lanes contain cleaved products from other Tay-Sachs patients. Lanes 15 and 16 contain cDNA from the patient of lane 14 but without hydroxylamine or piperidine, showing the specificity of the cleavage. c Diagram of the HEXA cDNA with the location of PCR primers for DGGE [the oligonucleotide primers used for PCR were 5′ GCCCGCCGTCCCGGCCCGACCCCCGCGCGTC CGGCGCCCGCGTCCTTTACCCGAACAACT 3′ (upstream primer with GC clamp) and 5′ CTGTGCTGTGTAGATGTGGG 3′ (downstream primer)] and CMC, and the location of 4 different mutations in patients BO, NA, DA and KM. The computerized analysis predicts a low-melting domain of 400 bp that is represented by a hatched bar in the 629-bp amplified fragment.
Methods
Cell lines The fibroblast cell lines came from Tay-Sachs patients diagnosed in our different European laboratories. Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with antibiotics and 10% fetal calf serum at 37°C in 5% CO2. β-Hexosaminidase A activity in fibroblasts was measured by a fluorometric assay using 4-methylumbelliferyl-β-D-N-acetylglucosaminide-6-sulfate as previously described [17]. All cell lines were totally deficient in β-hexosaminidase A activity with this artificial substrate. For each patient, 4 plates (65 cm2) were used to measure enzymatic activity and to extract total RNA 6751 and genomic DNA for mutation analysis.
CMC. This procedure was done according to Grompe et al. [19]. The labeling of the two strands of the PCR-amplified reference fragment was performed by a second amplification with radiolabeled primers as described [19]. The labeled probe (approximately 10 ng) was mixed with 15-fold molar excess of amplified target DNA in 0.3 M NaCl, 3.5 mM MgCl2, 3 mM Tris-HCl pH 7.7, boiled for 5 min, chilled on ice and then incubated at 42°C for 2 h. After hybridization, the samples were ethanol precipitated and resuspended in 20 µl water for further use.
Chemical Modification and Cleavage. The best results were obtained when heteroduplex DNA (6 µl for each reaction) was incubated either at 39°C for 1 h in 20 µl of a fresh 4 M solution of hydroxylamine hydrochloride (Aldrich) titrated to pH 6 by addition of diethylamine (Aldrich) for C-modification, or at 37°C for 1 h in 19 µl of a mix containing 10 mM Tris-HCl pH 8, 1 mM EDTA, 1.5% (v/v) pyridine and 0.025% osmium tetroxide [4% stock solution (Aldrich) stored at 4 °C] for T-modification. After piperidine cleavage (1 M fresh solution, 90°C for 30 min), the fragments were analyzed by electrophoresis in a 6% denaturing Polyacrylamide gel, followed by autoradiography [20].
DGGE. A melting map (plots of the midpoint melting temperature as a function of position along a DNA molecule) of the cDNA fragment with a GC clamp attached to the 5′ end was generated using the computer program of Lerman and Silverstein [21]. This program simulates the melting and mobility behavior of the clamped PCR fragment in the appropriate denaturing gradient gel. PCR-amplified fragments of cDNA were analyzed by DGGE in a Hoeffer SE 600 type apparatus (LKB) as previously described [22]. Samples (15 µl) were loaded onto a 6.5% Polyacrylamide gel (acrylamide/bisacrylamide = 37.5/1) containing a 30–80% denaturant gradient parallel to the direction of electrophoresis [100% denaturant = 7 M urea and 40% (v/v) formamide]. The gel was ran at 160 V for 3 h 30 min in TAE buffer maintained at a constant temperature of 60°C by a circulating heater and stirrer. After electrophoresis, the gel was stained with ethidium bromide.
Results and Discussion
Screening with ASOs for six known mutations showed marked differences in frequency (table 1). The 4-bp insertion in exon 11 (which represents 70% of Ashkenazi Jewish Tay-Sachs alleles) was found in our population with a frequency of 24% and is present in both Ashkenazi and non-Ashkenazi patients (see table 2 for ethnic origin). The French-Canadian mutation, accounting for 80% of alleles in Canada, was completely absent from our population suggesting that this mutation is rare in France/Europe or that it appeared in Canada after the French immigration [16].
The mutation described as responsible for the adult form [9, 10] of Tay-Sachs disease was present in 5% of alleles studied and represents 33% of alleles in our adult patients. We found the ΔPhe 304/305 mutation, first described in a Moroccan Jewish family [23], in 6% of alleles and consider it to be a panethnic mutation, since we identified it in French, Italian and Portuguese patients [20]. In the Moroccan Jewish population its frequency is about 25% of alleles.
Of particular interest is the IVS9*1 mutation identified in our laboratory [20], also reported in an English study [24] and by Akerman et al. [25], and characterized as a 17-bp insertion due to a GT to AT transition at the donor site of intron 9, resulting in the activation of a cryptic donor site in the intron [26]. This mutation seems to be frequent in the European population since it was found at 40% of alleles in a non-Jewish Tay-Sachs population from the British isles [24] and represents 11% of the alleles we studied.
Figure 1 shows comparative results between CMC and DGGE analysis of some of our patients with Tay-Sachs disease.
Mismatch analysis of the 5′ region of HEXA cDNA allowed us to detect 4 novel mutations, while DGGE analysis revealed the existence of melting polymorphisms for 7 out of 19 Tay-Sachs patients studied. For patients with only one identical band, we mixed normal and mutant sequences in a ratio of 1:1 to observe heteroduplexes that can easily distinguish between different mutations (fig. 1a).
Table 3 gives a general view of mutations characterized in the 42 Tay-Sachs patients studied. 28 different mutations were found in the 84 alleles, 20 of which were identified for the first time in this group of Tay-Sachs patients: 11 missense mutations, 2 nonsense mutations, 5 splicing mutations, and 2 frame-shift mutations.
Several general remarks can be made concerning the genotypes found in this Tay-Sachs population and the molecular mechanisms involved.
-
(1)
The substituted amino acids resulting from the missense mutations seem to play an important role since they occur in conserved positions as well as in the human HEXB, considered to be derived along with HEXA from a common ancestor gene, and in Dictyostelium discoideum HEXA [27]. Their substitution leads to a total loss of enzymatic activity.
-
(2)
Eight amino acid substitutions (Leu39 → Arg, Arg170 → Gln, Arg170 → Trp, Arg178 → His, Lys197 → Thr, His204 → Arg, Met301 → Arg, Glu482 → Lys) produce charge changes.
-
(3)
Arg499 → Cys can be responsible for changes in protein conformation [28].
-
(4)
The alternative splicing due to a G to A transversion at position −1 of exon 5 is a quantitative mutation, responsible for a strong decrease in donor site efficiency resulting in two low-abundance RNAs: one lacking exon 5 which is a nonenzyme producer, and the other normally spliced but carrying the silent G to A mutation and therefore coding for an absolutely normal enzyme reported to account for about 3% of residual HEXA activity in a patient with a late infantile form of Tay-Sachs disease [14].
-
(5)
The missense mutation (Leu39 → Arg) creates an aberrant peptidase site, Tyr-Val-Arg39, which might interfere with the HEXA maturation process [29].
-
(6)
Of particular interest for phenotypic correlations is the substitution of Arg499. When it is replaced by Cys, and in association with a null allele (4-bp insertion in exon 11), this mutation is responsible for an infantile form of the disease. When replaced by His and in association with the same null allele it was found in a juvenile form [30]. When replaced by His, but in association with the Lys197 → Thr substitution, the mutation is responsible for an adult Tay-Sachs form (table 2). We can conclude that Arg499 → His is a phenotypically mild allele and that Lys197→ Thr is a new adult allele.
-
(7)
40% of mutations occurred in a ‘hot spot’ CpG dinucleotide.
-
(8)
All mutations in the donor splicing site except one (IVS9*1) result in the excision of the previous exon and produce unstable mRNAs.
-
(9)
16 of the mutations we characterized seem to be very rare since we identified them on only one chromosome from 84 studied.
Table 2 shows the clinical form, ethnic origin and genotype found in the 42 patients studied and illustrates the molecular epidemiology of Tay-Sachs disease in a European population with various ethnic backgrounds. This study demonstrated once again the great heterogeneity of HEXA lesions involved in Tay-Sachs disease. Most of the patients studied are compound heterozygotes and it was found that the homozygotes are very often associated with consanguinity (table 2).
Most mutations are familial. The ethnic mutations — 4-bp insertion in exon 11, IVS9*1, ΔPhe304/305 — which were also the most frequent in our study, were found in patients of different ethnic origins, probably due to some degree of intermarriage.
Correlation of the genotype with the clinical form was possible in some cases. All patients with the B1 variant had the Arg178→ His mutation [31, 32] in a homozygous state. 4 patients with adult-type disease had the Gly269 → Ser mutation combined with different null alleles. In two other adult patients one had the Lys197 → Thr mutation combined with Arg499 → His, another the IVS6*1 mutation with an unidentified allele. Several patients with the infantile form had the 4-bp insertion in exon 11: 6 in a homozygous state and 6 in a heterozygous state combined with other different infantile mutations.
Our strategy allowed the characterization of 75 out of 84 alleles representing 90% of the total. The use of RNA as starting material can explain why 10% of the alleles were not identified since this technique is not suitable for detecting mutations in the promotor region; it also fails to detect some splicing mutations. Sequencing the promotor and all the intron/exon junctions can give complementary information.
Another limitation results from the disequilibrium between the number of transcripts corresponding to the two alleles in the case of compound heterozygotes. Genomic DNA analysis is necessary in this case.
In our experience, DGGE seems to be more efficient than CMC for screening for both known and unknown mutations in a limited segment of cDNA (400 bp). However, our data demonstrate that CMC is a valuable alternative for detecting nucleotide substitutions when the whole coding sequence of the HEXA gene must be analyzed.
Thus, an effective methodology is now available for the molecular characterization of genetic diseases. However, the extremely high molecular heterogeneity found in Tay-Sachs disease (46 different mutations have so far been identified) prevents the use of this methodology for current routine purposes, particularly when enzymatic methods are more simple, less expensive and sufficiently reliable.
Nevertheless, gene lesion characterization is important for a better understanding of pathogenesis, in some cases for the diagnosis of a particular phenotype, or to establish the prognosis, and with the goal of creating animal models with different phenotypical expressions necessary for the preliminary steps towards gene therapy.
References
Sandhoff K, Conzelmann E, Neufeld E, Kaback M, Suzuki K: The GM2 gangliosidoses; in Scriver CR, Beaudet AL, Sly WS, Valle D (eds): The Metabolic Basis of Inherited Disease, ed 6. New York, McGraw-Hill, 1989, pp 1807–1839.
Conzelmann E, Kytzia HJ, Navon R, Sandhoff K: Ganglioside GM2 N-acetyl-β-D-galactosaminidase activity in cultured fibroblasts of late-infantile and adult GM2 gangliosidosis patients and of healthy probands with low hexosaminidase level. Am J Hum Genet 1983;35:900–913.
Leinekugel P, Michel S, Conzelmann E, Sandhoff K: Quantitative correlation between the residual activity of β-hexosaminidase A and arylsulfatase A and the seventy of resulting lysosomal storage disease. Hum Genet 1992;88:513–523.
Arpaia E, Dumbrille-Ross A, Maler T, Neote K, Tropak M, Troxel C, Stirling JS, Bapat B, Lamhonwah AM, Mahuran DJ, Schuster SM, Clarke JTR, Lowden JA, Gravel RA: Identification of an altered splice site in Ashkenazi Tay-Sachs disease. Nature 1988;333:85–86.
Ohno K, Suzuki K: A splicing defect due to an exon-intron junctional mutation results in abnormal β-hexosaminidase a chain mRNAs in Ashkenazi Jewish patients with Tay-Sachs disease. Biochem Biophys Res Commun 1988;153:463–469.
Myerowitz R: Splice junction mutation in some Ashkenazi Jews with Tay-Sachs disease: Evidence against a single defect within this ethnic group. Proc Natl Acad Sci USA 1988;85:3955–3959.
Myerowitz R, Costigan C: The major defect in Ashkenazi Jews with Tay-Sachs disease is an insertion in the gene for the α-chain of β-hexosaminidase. J Biol Chem 1988;263:18587–18589.
Paw BH, Tieu PT, Kaback MM, Lim J, Neufeld EF: Frequency of three Hex A mutant alleles among Jewish an non-Jewish carriers identified in a Tay-Sachs screening program. Am J Hum Genet 1990;47:698–705.
Paw BM, Kaback MM, Neufeld EF: Molecular basis of adult-onset and chronic GM2 gangliosidosis in patients of Ashkenazi Jewish origin: Substitution of serine for glycine at position 269 of the α-subunit of β-hexosaminidase. Proc Natl Acad Sci USA 1989;86:2413–2417.
Navon R, Proia RL: The mutation in Ashkenazi Jews with adult GM2 gangliosidosis, the adult form of Tay-Sachs disease. Science 1989; 243:1471–1474.
Myerowitz R, Hogikyan ND: A deletion involving Alu sequences in the β-hexosaminidase α-chain gene of French Canadians with Tay-Sachs disease. J Biol Chem 1987;262:15386–15399.
Hechtman P, Kaplan F, Bayleran J, Boulay B, Andermann E, de Braekeleer M, Melancon S, Lambert M, Potier M, Gagné R, Kolodny E, Clow C, Capua A, Prevost C, Scriver C: More than one mutant allele causes infantile Tay-Sachs disease in French-Canadians. Am J Hum Genet 1990:47:815–822.
Gravel RA, Triggs-Raine BL, Mahuran DJ: Biochemistry and genetics of Tay-Sachs disease. Can J Neurol Sci 1991;18(suppl 3):419–423.
Akli S, Chelly J, Mezard C, Gandy S, Kahn A, Poenaru L: A ‘G’ to ‘A’ mutation at position −1 of a 5′ splice site in a late infantile form of Tay-Sachs disease. J Biol Chem 1990;265:7324–7330.
Cotton RGH, Rodrigues NR, Campbell RD: Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci USA 1988;85:4397–4401.
Kaplan F, Boulay B, Bayleran J, Hechtman P: Allele specific amplification of genomic DNA for detection of deletion mutations: Identification of a French Canadian Tay-Sachs mutation. J Inherit Metab Dis, in press.
Bayleran J, Hechtman P, Saray W: Synthesis of 4-MU-β-D-N-acetylglucosamine-6-sulfate and its use in classification of GM2 gangliosidosis genotypes. Clin Chim Acta 1984;143:73–89.
Chirgwin JM, Przybyla AE, Macdonald RJ, Rutter WJ: Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 1979;18:5294–5299.
Grompe M, Muzny DM, Caskey CT: Scanning detection of mutations in human ornithine transcarbamoylase by chemical mismatch cleavage. Proc Natl Acad Sci USA 1989;86:5888–5892.
Akli S, Chelly J, Lacorte JM, Poenaru L, Kahn A: Seven novel Tay-Sachs mutations detected by chemical mismatch cleavage of PCR-amplified cDNA fragments. Genomics 1991;11:124–134.
Lerman LS, Silverstein K: Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol 1987;155:482–501.
Myers RM, Maniatis T, Lerman LS: Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol 1987;155:501–527.
Drucker L, Proia RL, Navon R: Identification and rapid detection of three Tay-Sachs mutations in the Moroccan Jewish population. Am J Hum Genet 1992;51:371–377.
Landels EC, Green PM, Ellis IH, Fensom AH, Bobrow M: Beta-Hexosaminidase splice site mutation has a high frequency among non-Jewish Tay-Sachs disease carriers from the British Isles. J Med Genet 1992;29:563–567.
Akerman BR, Zielenski J, Triggs-Raine BL, Prence EM, Natowicz MR, Lim-Steele JST, Kaback MM, Mules EH, Thomas GH, Clarke JTR, Gravel RA: A mutation common in non-Jewish Tay-Sachs disease: Frequency and RNA studies. Hum Mutat 1992;1:303–309.
Akli S, Chelly J, Kahn A, Poenaru L: A null allele frequent in non-Jewish Tay-Sachs patients. Hum Genet 1993;90:614–620.
Graham TR, Zassenhaus HP, Kaplan A: Molecular cloning of the cDNA which encodes β-N-acetylhexosaminidase A from Dictyostelium discoideum. J Biol Chem 1988;263:16823–16829.
Mules EH, Hayflick Z, Miller CS, Reynolds LW, Thomas GH: Six novel deleterious and three neutral mutations in the gene encoding the a subunit of Hex A in non-Jewish individuals. Am J Hum Genet 1992;50:834–841.
Akli S, Chomel JC, Lacorte JM, Bachner L, Kahn A, Poenaru L: Ten novel mutations in the HEXA gene in non-Jewish Tay-Sachs patients. Hum Mol Genet 1993;2:61–67.
Paw BH, Moskowitz SM, Urhammer N, Wright N, Kaback MM, Neufeld EF: Juvenile GM2 gangliosidosis caused by substitution of histidine for arginine at position 499 or 504 of the α-subunit of β-hexosaminidase. J Biol Chem 1990;265:9452–9457.
Tanaka A, Ohno K, Sandhoff K, Maire I, Kolodny EH, Brown A, Suzuki k: GM2-gangliosidosis B1 variant: Analysis of β-hexosaminidase α gene abnormalities in seven patients. Am J Hum Genet 1990;27:465–473.
Nakano T, Muscillo M, Ohno K, Suzuki Y, Suzuki K: A new point mutation within exon 5 of β-hexosaminidase α gene in a Japanese infant with Tay-Sachs disease. J Neurochem 1988;51:984–987.
Acknowledgements
We thank Dr. L. Lerman for kindly making the computer program available and Mrs Ajroldi for typing this manuscript. This work was supported by grants from A.F.M. (Association Française contre les Myopathies) and V.M.L. (Vaincre les Maladies Lysosomales).
Author information
Authors and Affiliations
Additional information
This work was coordinated at the Institut Cochin de Génétique Moléculaire, Paris, France.
Rights and permissions
About this article
Cite this article
Akli, S., Boue, J., Sandhoff, K. et al. Collaborative Study of the Molecular Epidemiology of Tay-Sachs Disease in Europe. Eur J Hum Genet 1, 229–238 (1993). https://doi.org/10.1159/000472416
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472416
Key Words
This article is cited by
-
Identification of novel variants in a large cohort of children with Tay–Sachs disease: An initiative of a multicentric task force on lysosomal storage disorders by Government of India
Journal of Human Genetics (2019)
-
Disruption of murine Hexa gene leads to enzymatic deficiency and to neuronal lysosomal storage, similar to that observed in Tay-Sachs disease
Mammalian Genome (1995)