Abstract
A deletion hybrid breakpoint map of the chromosomal region 13q14–q21 has been constructed using 19 DNA markers and 13 cell lines with breakpoints in this chromosomal region. The cell lines define 10 distinct intervals in this region, which spans approximately 20 Mb. The markers include 6 RFLP markers, 11 microsatellites that provisionally had been mapped to the region 13q14–21, and 2 new polymorphic CA-repeats that were developed from an EMBL3 library of cell line ICD, containing 13pter-q14.3. The following order of markers was established: CEN — D13S320 — (D13S118, D13S153) — RB1 — D13S319 — D13S25 — (D13S31, D13S59, D13S133, D13S137) — D13S163 — D13S119 — (D13S26, D13S55) — (D13S131, D13S134, D13S135, D13S144, D13S152) — TEL.
Similar content being viewed by others
Introduction
Several disease genes of medical importance have been assigned to the region 13q14-q21 of chromosome 13. Deletions in the region, occurring in a proportion of retinoblastoma patients, provided a clue to the identification and isolation of the RB1 gene [1]. Other chromosomal rearrangements in the region found in association with chronic B-cell leukaemia, indicate that it may be the location of a gene involved in the development of this form of leukaemia [2]. Linkage analysis has shown that the locus for Wilson disease (WND), an autosomal recessive disorder of copper metabolism, is tightly linked to the chromosomal marker D13S31 [3, 4] positioned at the junction of chromosomal bands 13q14.3-q21.1 [5]. Recent abstracts suggest that the gene has been cloned [6, 7].
In order to identify microsatellite markers in the 13q14 region, two CA-repeats were isolated from clones from a lambda phage library from hybrid cell line ICD, containing 13pter-q14.3 as its sole human material [8], that also hybridized to cell line E8 (13q14.1-qter) as detected by fluorescent in situ hybridization (FISH) [9].
These CA-repeats were mapped on a panel of 13 cell lines with breakpoints in the chromosomal region 13q 14–21. In addition, we mapped 6 restriction fragment length polymorphism (RFLP) [3, 4, 10, 13], and 11 microsatellites from three different genetic maps [10–12] on this hybrid panel. The three genetic maps provide only approximate locations of the loci, the map of Bowcock et al. [10] being the only one which also includes the RFLP markers previously analyzed. Unfortunately, these three maps have no markers in common except for two loci at 13q32-q34, D13S64 and D13S71, which appear on the maps of Bowcock et al. [10] and Petrukhin et al. [11].
In the present study, we present a deletion hybrid breakpoint map of the chromosomal region 13q14–21, divided in 10 distinct intervals, which integrates the relative positions of the markers from the three genetic maps [10–12], and from the earlier RFLP map [13], and of 2 newly isolated microsatellites.
Materials and Methods
Hybrid Cell Lines
Hybrid cell lines GF7 (human chromosome 13 only [8]), ICD (13pter-13q14.3 [8]), WC-H38B3B6 (13pter-q13::13q21.1 -qter [14]), WC-H12D12 (13pter-q14::13q22-qter [14]), NM-87-26XT (13q12-q14:: 13q22-qter [15]), PKII-90-P5b (= PK88-25, 13pter-q12::13q21.2-qter [15]), KSF39 (13pter-q14.1 [16]), KBF11 (13pter-q12::13q14-qter [16]), E8 (13q14.1-qter[17]), D1 (13pter-q14.1 [17]), RHF 407 (13q14.3–13qter [2]), RHF 2324 (13pter-13q14.3 [2]), GS89a (13pter-q14::13q32–34 [D. Warburton, unpubl.]), and CF27 (13pter-q14.1::13q22-qter [T.K. Mohandas, unpubl.] were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum in the presence of antibiotics. All hybrid cell lines except GF7 and ICD contain other human chromosomes in addition to the der(13). DNA was isolated after treatment of the cells with SDS and proteinase K by phenol extraction according to standard procedures.
Plasmid Cloning, Sequencing and Primer Design
(CA)n-containing EMBL3 clones λ42.6 and λ42.7 were isolated from a phage lambda library from cell line ICD [8]. Phage λ42.6 was cut with Sau3A and ligated in the BamHI site of pBluescriptKS(−). Phage λ42.1 was cut with RsaI, HaeIII, and AluI and ligated in the EcoRV site of pBluescriptKS(−). The ligation mix was transformed into Escherichia coli JM83 or DH5α competent cells, respectively. (CA)n-containing subclones were identified by colony hybridization with an end-labelled (GT)12 oligonucleotide. The clones were sequenced from both ends using the T7 sequenase kit (Pharmacia LKB, Uppsala, Sweden) and standard primers. Primers on both sides of the CA-repeats were selected with the aid of the computer program ‘primer designer’ (Scientific & Educational Software, Need City, Pa., USA).
PCR Reactions
Reactions were carried out in 20-µl volumes of ‘Supertaq reaction buffer’ (Sphearo Q HT Biotechnology, Leiden, The Netherlands) with final concentrations of 10mM Tris-HCl (pH = 9.0), 50 mM KCl, 1.5 mM Mg2+, 0.1% Triton X-100, 0.01% gelatin, and including 0.2 mM of each dNTP (dATP, dCTP, dGTP, dTTP), 0.125 U Taq polymerase (Supertaq, Sphearo Q HT Biotechnology), 100 ng of each primer and 100–300 ng template DNA. After an initial denaturation step of 93° C for 2 min, thermal cycling was carried out for 27 cycles consisting of denaturation at 93°C for 1 min, annealing at 5–10°C below Td for 1 min, and extension at 72° C for 1 min, followed by a final extension step at 72 °C for 5 min. For some primer sets, it was necessary to increase the annealing temperature of the first cycles to increase specificity. PCR products were visualized on agarose gels, stained with ethidium bromide.
Radioactive PCR was carried out by substituting 1/10 of the dCTP by [α-32P]dCTP. Products were size separated on 4–6% Polyacrylamide gels and analyzed after exposure to an X-ray-sensitive film.
Results
Development of New Microsatellite Markers
Two human insert-containing phages from an EMBL3 library from cell line ICD [9], shown by FISH to lie distal to the translocation breakpoint of the der(13) involved in alveolar rhabdomyosarcoma in cell lines E8 and D1, contained a CA-repeat as determined by hybridization with a (GT)12 oligonucleotide. (CA)n-containing fragments of these phages, designated λ42.6 and λ 42.7, were sub-cloned and sequenced (see Materials and Methods). The designed primers flanking the CA-repeat are presented in table 1. Both CA-repeats were polymorphic with heterozygosities of 41 and 82%, respectively.
Order of Markers
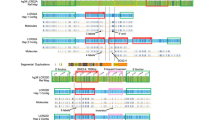
The 13 der(13) hybrid cell lines (see Materials and Methods) were analyzed by PCR for the presence or absence of the RFLP markers RB1, D13S25, D13S26, D13S31, D13S55, D13S59, and the microsatellite sequences D13S118, D13S119, D13S131, D13S133, D13S134, D13S135, D13S137, D13S144, D13S152, D13S153, D13S163, D13S319, D13S320. Primer source and sequences are indicated in table 2. The cell lines divide the region 13q14-q21 into 10 distinct intervals (fig. 1). The order of markers was: CEN — D13S320 — (D13S118, D13S153) — RB1 — D13S319 — D13S25 — (D13S31, D13S59, D13S133, D13S137) — D13S163 — D13S119 — (D13S26, D13S55) — (D13S131, D13S134, D13S135, D13S144, D13S152) — TEL. All hybrids were tested for all markers. No gaps were identified in the deletion hybrids. All markers were present on the monochromosomal cell line GF7.
a A deletion hybrid breakpoint map of the region 13q14–q21. The der(13) are indicated by horizontal bars. Equal length has arbitrarily been attributed to all intervals. The putative position of the WND gene is indicated by a black arrow, that of the BCLL gene by the spotted arrow, b Breakpoints in 13q14–q21 from the der(13) of the cell lines used in this study. Modified after Bowcock and Taggart [23].
Discussion
The relative order of the microsatellite markers is in agreement with the order that the subsets have on each of the genetic maps [10–12]. This also applies to the relative order of the RFLP markers [3, 13]. The order along the chromosome as determined by linkage analysis is thus confirmed using physical mapping.
The cell hybrids used define 10 distinct intervals in the PCR-based hybrid breakpoint map of the chromosomal region 13q14-q21. The map stretches from the translocation breakpoint of the der(13) of cell lines E8 and D1 in 13q14.1 to the proximal breakpoint of the distal segment of the der(13) of cell line PKII-90-P5b at 13q21.2, an estimated distance of 20 Mb [derived from 18].
The smallest deletion seen in BCLL is determined by the cell lines RHF 407 and RHF 2324, which contain the different translocation products from a translocation involving 13q14 in a B-cell chronic lymphocytic leukaemia patient [2]. Therefore, BCLL potentially maps in interval 4 or 5 of the map. WND is closely linked to the marker D13S31 [3, 4], and between the markers D13S31 and D13S59 [3]. As D13S31 and D13S59 are situated in interval 6, WND will also be situated in this interval, which must be in proximal 13q21.1 [5].
References
Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP: A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986;323:643–646
Hawthorn LA, Chapman R, Oscier D, Cowell JK: The consistent 13q14 translocation breakpoint seen in chronic B-cell leukaemia involves a deletion of the D13S25 locus which lies distal to the retinoblastoma predisposition gene. Oncogene 1993;8:1415–1419
Farrer LA, Bowcock AM, Herbert JM, Bonné-Tamir B, Sternlieb I, Giagheddu M, St George-Hyslop P, Frydman M, Lössner J, Demelia L, Carcassi C, Lee R, Beker R, Bale AE, Donis-Keller H, Scheinberg IH, Cavalli-Sforza LL: Predictive testing for Wilson’s disease using tightly linked and flanking DNA markers. Neurology 1991;41:992–999
Scheffer H, Houwen RHJ, Te Meerman GJ, Loessner J, Bachmann B, Kunert E, Verlind E, Buys CHCM: Identification of crossovers in Wilson disease families as reference points for a genetic localization of the gene. Hum Genet 1992;89:607–611
Kooy RF, Van der Veen AY, Verlind E, Houwen RHJ, Scheffer H, Buys CHCM: Physical localisation of the chromosomal marker D13S31 places the Wilson disease locus at the junction of bands q14.3 and q21.1 of chromosome 13. Hum Genet 1993;91:504–506
Tanzi RE, Petruhkin K, Soares B, Wasco W, Romano D, Ross B, Chernov I, Pirastu M, Hyslop PG, Gusella JF, Scheinberg IH, Gilliam TC: Isolation and characterization of a candidate gene for Wilson’s disease. Am J Hum Genet 1993; 53(suppl):228.
Cox DW, Bull PC, Rommens JM, Thomas GR: Physical mapping and haplotype studies define the region of the Wilson disease locus. Human Genome Mapping Workshop, Kobe 1993, 13–5.
Buys CHCM, Scheffer H, Perason PL, Aanstoot GH, Goor N, Niehaus AJ: Construction and application of a hybrid cell panel for regional localization of chromosome 13-specific DNA probes. Cytogenet Cell Genet 1985;40:597.
Shapiro DN, Sublett JE, Btau L, Valentine MB, Johnson CV, McNeil JA, Lawrence JB: Regional localization of 35 anonymous DNA probes relative to the 13q14 breakpoint in alveolar rhabdomyosarcoma by fluorescence in situ hybridization. Cytogenet Cell Genet 1993;62:105–106
Bowcock A, Osborne-Lawrence S, Barnes R, Chakravarti A, Washington S, Dunn C: Microsatellite polymorphism linkage map of human chromosome 13q. Genomics 1993;15:376–386
Petrukhin KE, Speer MC, Cayanis E, De Fátima Bonaldo M, Tantravahi U, Bento Soares M, Fischer SG, Warburton D, Gilliam TC, Ott J: A microsatellite genetic linkage map of human chromosome 13. Genomics 1993;15:76–85
Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, Lathrop M: A second generation linkage map of the human genome. Nature 1992;359:794–801
Bowcock AM, Farrer LA, Herbert JA, Bale AE, Cavalli-Sforza L: A contiguous linkage map of chromosome 13q with 39 distinct loci separated on average by 5.1 centimor-gans. Genomics 1991;11:517–529
Griffin CA, Emanuel BS, Hansen JR, Cavenee WK, Myers JC: Human collagen genes encoding basement membrane αl(IV) and α2(IV) chains map to the distal long arm of chromosome 13. Proc Natl Acad Sci USA 1987;84:512–516
Warburton D, Gersen S, Yu M-T, Jackson C, Handelin B, Housman D: Monochromosomal rodent-human hybrids from microcell fusion of human lymphoblastoid cells containing an inserted dominant selectable marker. Genomics 1990;6:358–366
Cowell JK, Mitchel CD: A somatic cell hybrid mapping panel for regional assignment of human chromosome 13 DNA sequences. Cytogenet Cell Genet 1989;52:1–6
Shapiro DN, Valentine M, Sublett J, Tereba A, Scheffer H, Buys CHCM, Look AT: Subchromosomal localization of the t(2,13) breakpoint in alveolar rhabdomyosarcoma. Genes Chromosomes Cancer 1992;4:241–249
Morton NE: Parameters of the human genome. Proc Natl Acad Sci USA 1991;88:7474–7476
Vaughn GL, Toguchida J, McGee TL, Dryja TP: PCR detection of the Tth111I RFLP at the RB locus. Nucleic Acids Res 1990;18:4965.
Stuart EA, White A, Tomfohrde J, Osborne-Lawrence S, Prestridge L, Bonne-Tamir B, Scheinberg IH, St George-Hyslop P, Giagheddu M, Kim J-W, Seo JK, Lo WH-Y, Ivanova-Smolenskaya IA, Limborska SA, Cavalli-Sforza LL, Farrer LA, Bowcock AM: Polymorphic micro-satellites and Wilson disease. Am J Hum Genet 1993;53:864–873
Lalande M, Dryja TP, Schreck RR, Shipley J, Flint A, Latt SA: Isolation of human chromosome 13-specific DNA sequences cloned from flow-sorted chromosomes and potentially linked to the retinoblastoma locus. Cancer Genet Cytogenet 1984;13:283–295
Donis-Keller H, Green P, Helms C, Cartinhour S, Weiffenbach B, Stephens K, Keith TP, Bowden DW, Smith DR, Lander ES, Botstein D, Akots G, Rediker KS, Gravius T, Brown VA, Rising MB, Parker C, Powers JA, Watt DE, Kauffman ER, Bricker A, Philipps P, Muller-Kahle H, Fulton TR, Ng S, Schummm JW, Braman JC, Knowlton RG, Barker DF, Crooks SM, Linkoln SE, Daly MJ, Abrahamson J: A genetic linkage map of the human genome. Cell 1987;51:319–337
Bowcock AM, Taggart RT: Report of the committee on the genetic constitution of chromosome 13. Human gene mapping 11. Cytogenet Cell Genet 1991;58:580–604
Acknowledgements
We gratefully acknowledge Dr. W.K. Cavenee for making available cell lines WC-H38B3B6 and WCH12D12, Dr. D. Warburton for cell lines NM-87-26XT, PKII-90-P5b, and GS89a, and Drs. T.K. Mohandas and A. Chakravarti for cell line CF27. This work was supported by the Netherlands Organisation for Scientific Research.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kooy, R.F., Verlind, E., Houwen, R.H.J. et al. A Deletion Hybrid Breakpoint Map of the Chromosomal Region 13q14–q21 Orders 19 Genetic Markers in 10 Intervals. Eur J Hum Genet 2, 59–65 (1994). https://doi.org/10.1159/000472342
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472342