Abstract
L1 is a neuronal cell adhesion molecule with important functions in the development of the nervous system. The gene encoding L1 is located near the telomere of the long arm of the X chromosome in Xq28. We review here the evidence that several X-linked mental retardation syndromes including X-linked hydrocephalus (HSAS), MASA syndrome, X-linked complicated spastic paraparesis (SP1) and X-linked corpus callosum agenesis (ACC) are all due to mutations in the L1 gene. The inter- and intrafamilial variability in families with an L1 mutation is very wide, and patients with HSAS, MASA, SP1 and ACC can be present within the same family. Therefore, we propose here to refer to this clinical syndrome with the acronym CRASH, for Corpus callosum hypoplasia, Retardation, Adducted thumbs, Spastic paraplegia and Hydrocephalus.
Similar content being viewed by others
Clinical Aspects
X-Linked Hydrocephalus
X-linked hydrocephalus (MIM No. 307000) was originally described by Bickers and Adams in 1949. It is referred to as HSAS for Hydrocephalus due to Stenosis of the Aqueduct of Sylvius. This designation was based upon the presence of aqueductal stenosis in many HSAS patients [1]. However, later studies reported several HSAS patients with hydrocephalus but without aqueductal stenosis [2]. Therefore, the occurrence of aqueductal stenosis in X-linked hydrocephalus might not be the cause of hydrocephalus. Nevertheless, the designation of HSAS is still used in the literature. HSAS is the most common genetic cause of hydrocephalus with an incidence of approximately 1/30,000 male births, and it is responsible for about one quarter of male patients with hydrocephalus not associated with spina bifida [3]. The clinical spectrum of HSAS is very broad and shows great intra- and interfamilial variation. The only obligate feature is mental retardation, with IQs usually ranging between 20 and 50. Hydrocephalus presents from impressive macrocephaly, resulting in pre- or perinatal death, to minimal enlargement of the ventricles. Moreover, HSAS families have been described in which some mentally retarded members did not have hydrocephalus [4, 5]. Adducted thumbs are seen in approximately 90% of the cases. Other features are spastic paraplegia, nystagmus and structural brain defects such as hypoplasia of the corpus callosum and septum pellucidum [5]. HSAS is a recessive X-linked disorder with no symptoms in carrier females. In 1990, we assigned the HSAS disease locus to the distal part of the long arm of the X chromosome in Xq28 by linkage analysis [6]. Further mapping studies refined the disease locus to a region of 2 Mb, distal to DXS52 and proximal to F8C [7, 8]. As the neural cell adhesion molecule L1 (L1CAM) is located in this interval, we suggested that HSAS might be due to mutations in L1 [7].
MASA Syndrome
In 1974, Bianchine and Lewis described an X-linked disorder characterized by Mental retardation, Aphasia, Shuffling gait and Adducted thumbs [9]. Referring to these four clinical features, the name ‘MASA syndrome’ was assigned to this disease (MIM No. 303350). Several MASA families have been reported, and some families have been redefined as representative of the MASA syndrome [9–17]. The clinical features show inter- and intrafamilial variation, with mental retardation being the only obligatory symptom. Although MASA syndrome is an X-linked recessive condition, some expressing carrier females have been reported, possibly due to unfavorable lyonization of the wild-type gene [9,14]. Corpus callosum agenesis or dysgenesis is also a frequent symptom in MASA patients [15]. Even though earlier studies [9] noticed the clinical overlap between MASA and HSAS, a clear distinction was made between those two conditions, based upon the presence or absence of hydrocephalus in HSAS and MASA, respectively. However, in later clinical studies, the sharp boundaries between HSAS and MASA began to fade away as (1) HSAS families were reported in which some affected members did not have hydrocephalus [4, 5], (2) some MASA families contained HSAS patients with hydrocephalus [16], and (3) linkage studies assigned the MASA disease locus to Xq28, the same region as HSAS [16, 17]. This led us to suggest that HSAS and MASA were allelic disorders due to mutations in the same gene [6].
Spastic Paraplegia
Spastic Paraplegia (SP) is a clinically and genetically heterogeneous condition. Some investigators divide SP into pure and complicated forms [18]. In pure SP, spasticity of the lower extremities is the only clinical sign. In complicated SP, additional symptoms such as mental retardation and/or congenital brain malformations are present. Most hereditary forms of SP are inherited in an autosomal dominant or recessive way, whereas X-linked SP is rare. A locus for X-linked SP (SP2) has been localized on the long arm of the X chromosome in Xq21-q22, both in families with pure [19] and complicated [20] SP. This latter form of SP results from mutations in the proteolipid protein (PLP) gene and is allelic to Pelizaeus-Merzbacher disease (MIM No. 312080), a severe X-linked disorder in which progressive demyelination of the central nervous system occurs [21]. Kenwrick et al. [12] have described a form of X-linked complicated SP (SP1, MIM No. 312900) in a family with SP, mental retardation and adducted thumbs. Linkage analysis assigned the SP1 disease locus to the distal part of the long arm of the X chromosome in Xq28. Later clinical studies of this SP1 family revealed that the clinical spectrum was compatible with MASA syndrome [17], suggesting that SP1 and MASA syndrome are coallelic.
Agenesis of the Corpus Callosum
The corpus callosum is the largest fiber tract in the brain, and plays a key role in the communication between the two brain hemispheres. Corpus callosum agenesis (ACC) or dysgenesis (DCC) is a rather aspecific abnormality. Many cases of DCC, and to a lesser extent ACC, are found incidentally at necropsy in individuals who had normal cerebral function. The frequency of ACC/DCC in the general population has been estimated at 0.3–0.7%, whereas it is found in 2–3% of the mentally retarded. In most cases, ACC/DCC is an isolated finding. However, ACC and DCC can also occur as part of a syndrome, when accompanied by a number of other malformations. Many chromosomal and monogenic disorders have ACC/DCC in their spectrum of anomalies. X-linked forms of ACC/DCC, however, are very rare. Aicardi syndrome, an X-linked disorder that occurs only in females, has a clinical spectrum including ACC, chorioretinal lacunae, infantile spasms and psychomotor retardation. The gene involved in Aicardi syndrome has provisionally been mapped to Xp22. FG syndrome is an X-linked recessive condition with ACC and a variety of symptoms including macrocephaly, anal anomalies, Hirschsprung’s disease, constipation, hypotonia and mental retardation [22]. The gene responsible for FG syndrome has been mapped to Xq13 [Briault, pers. commun.]. Several ACC/DCC syndromes with unknown sublocalization on the X chromosome have been described. Menkes et al. [23] have reported a family with X-linked ACC/DCC, seizures, mental retardation and Hirschsprung’s disease, which might represent an example of FG syndrome. An X-linked DCC family with hydrocephalus, microcephaly, mental retardation and brachydactyly has been reported by Kang et al. [24]. Kaplan [25] reported two males with complete absence of the corpus callosum and severe psychomotor retardation, one of the patients having adducted thumbs and slightly enlarged ventricles. This spectrum of anomalies is reminiscent of MASA syndrome. The fact that ACC/DCC in some families might represent MASA, is also corroborated by the finding that ACC or DCC have been reported in patients with MASA syndrome [15] and HSAS [5]. This suggests that there might be just one gene for HSAS, MASA, SP1 and ACC/DCC.
Molecular Aspects
Identification of L1 as the Disease-Causing Gene in HSAS, MASA, SP1 and ACC/DCC
The localization of the HSAS gene in Xq28 in a 2-Mb interval containing the L1 (L1CAM) gene suggested that L1 was a candidate gene for HSAS [7]. Therefore, extensive mutation analysis was performed in the L1 gene from a large number of HSAS families. In 1992, the first L1 mutation in an HSAS family was reported by Rosenthal et al. [26]. This was a novel mutation in a branch point signal of an intron, leading to aberrant splicing. Soon thereafter, we discovered a second LI mutation [27], a duplication in the original HSAS family, which was used to localize the HSAS gene [5]. This confirmed that mutations in the LI gene are responsible for HSAS. Since then, we and others have found many LI mutations in HSAS families [28–30; Ken-wrick and Willems, unpubl. results].
As MASA syndrome was thought to be allelic with HSAS, the L1 gene of MASA patients was then analyzed, and several L1 mutations were found [30–32]. L1 mutations were also found in the original complicated SP1 family [30] which was later redefined as a MASA family [12], and in an ACC/DCC family with MASA features [15, 32]. This proves that HSAS, MASA, SP1 and ACC/DCC are allelic disorders due to mutations in a single gene, L1 (table 1).
L1 Protein and Function
L1, also referred to as L1 CAM, is a neuronal cell adhesion molecule belonging to the superfamily of the immunoglobulins [33, 34]. It is a transmembrane protein, expressed on the outgrowing axon of postmitotic nerve cells both in the central and peripheral nervous system and on Schwann cells. L1 is implicated in the pathfinding of an outgrowing axon and in neural cell migration during the development of the nerve system, as it plays a role in the adhesion between neurons, stimulates neurite outgrowth and fasciculation, and mediates the interaction between neurons and Schwann cells during meylination [35–38]. L1 is also important in the regeneration of damaged nerve tissue [39].
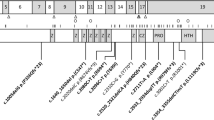
The protein has a molecular mass of 200 kD with 138 kD due to polypeptide and the rest to carbohydrate [40]. It is composed of various domains including (1) a cytoplasmic domain which is highly conserved among various species, (2) a transmembrane domain, composed of 23 hydrophobic residues, (3) five fibronectin-III-like domains, and (4) six immunoglobulin-type C2-like domains, each containing one disulfide bridge (fig. 1, 2). The extracellular part of L1 interacts with its surroundings through homophilic binding with other L1 molecules [41] and through heterophilic interactions with other neural cell adhesion molecules such as axonin-1/TAG-1, F3/F11 and phosphocan. The cytoplasmic tail of L1 interacts with ankyrin, a protein involved in the binding of the cytoskeleton to the cell membrane. It is unclear whether the cytoplasmic domain of L1 only offers an anchor to the cytoskeleton, or is also part of a secondary signal transduction system (fig. 2a). The different effects of L1 are not only mediated by cell adhesion, but are also the consequence of intracellular changes brought about by binding of LI to the fibroblast growth factor receptor (FGFR). Recently, evidence has been provided that L1 activates the FGFR tyrosine kinase upon ligand binding [42] (fig. 2a). The activated FGFR then phosphorylates phospholipase C, which triggers a cascade of events producing arachidonic acid. The elevated concentration of arachidonic acid causes a Ca2+ influx into the cell through N- and L-type Ca2+ channels. The same cascade is activated by soluble forms of the L1 molecule, suggesting that cell adhesion as such is not required to activate this signal transduction pathway [43]. Four different FGFR genes have been identified. All have a motif of five consecutive amino acids homologous to a protein sequence located between the third and fourth immunoglobulin domains of L1. In the same FGFR domain amino acid stretches are found which show homology to N-CAM and members of the Cadherin family. This domain is now referred to as the FGFR CAM-homology domain (FGFR-CHD).
Hypothetical models for the disturbance of normal L1 function by different mutations, indicated by an asterisk, a Normal functioning of L1. Hemophilic interaction between L1 molecules on neighboring neurons. The fibroblast growth factor receptor (FGFR) interacts with L1 via its CAM homology domain and dimerizes upon activation. This leads to activation of the two tyrosine kinase domains, b A missense mutation in the extracellular part of L1. The homophilic interaction between L1 molecules is weakened or absent, leading to absent or abnormal intracellular transduction, c A mutation in the cytoplasmic domain. Adhesion between neurons, and between L1 and the FGFR remains unaffected as the extracellular domains are intact. However, the cytoplasmic domain is truncated or absent. This might lead to absent or abnormal intracellular transduction, or impaired interaction with the cytoskeleton. d A nonsense mutation in one of the immunoglobulin domains leading to a premature extracellular stop codon. As the truncated protein lacks some immunoglobulin domains, the fibronectin domains, the transmembrane segment and the intracellular domain, it is secreted into the extracellular space. There it might be rapidly degraded, it might be stable but not functional, or it might block normal interaction of other cell adhesion molecules with immunoglobulin domains.
The L1 Gene
The L1 gene spans 16 kb [Rosenthal, pers. commun.] and is composed of 28 exons. The open reading frame has 3,825 bp, and encodes a protein of 1,275 amino acids. The first 19 amino acids form a signal peptide that is cleaved from the native protein, which is 1,256 amino acids long. The exons 3–14 encode the 6 immunoglobulin domains with two exons for each domain (fig. 1). Exons 15–24 encode the 5 fibronectin domains, exon 25 the transmembrane domain and exons 26–28 form the cytoplasmatic domain. An alternative splicing variant lacking four amino acids in the cytoplasmatic domain has been observed in the peripheral nerve system [44] but its functional importance is unknown. Structural and functional homologues of L1 have been found in mouse (L1), rat (NILE), chick (G4/8D9) and Drosophila (neuroglian). The cytoplasmic domain is by far the most conserved, sharing 100% homology at the amino acid level between human and mouse [34].
L1 Mutations
All mutations found to date are private mutations that have never been observed in more than one family. The mutations are dispersed throughout the whole L1 gene, and include deletions, a duplication, missense, nonsense, splice site mutations, and a novel type of mutation in a branch point signal leading to aberrant splicing. An overview of all L1 mutations reported so far in HSAS, MASA, SP1 and ACC/DCC is given in table 1.
The mutations can be classified into three broad categories. The first class consists of missense point mutations in the extracellular domain (mutations 2–5, 7). In these cases, the mutant protein is still an integral membrane protein with a normal transmembrane and cytoplasmic domain. However, the binding capacities of the L1 protein towards its ligands or its signaling pathways might be distorted, as the structure of the extracellular domains has been altered at one amino acid position (fig. 2b). The second class of mutations has a disrupted cytoplasmic domain, either by missense mutation (mutation 11), frameshift (mutation 10) or large rearrangements such as a duplication (mutation 12) or a deletion (mutation 13). In all of these cases, the extracellular and transmembrane domains remain intact (fig. 2c). The influence of these mutations on the functioning of L1 is unclear. It is possible that mutations in the cytoplasmic domain disturb the transduction of extracellular signals into the cell via an unknown signal pathway. Alternatively, it is possible that these mutations lead to impaired binding with the intracellular cytoskeleton. The third class of mutant L1 proteins has a premature stop codon in the extracellular part, due to a nonsense mutation (mutation 6) or a frameshift mutation (mutations 1 and 9). The mutant protein could be secreted into the extracellular space as no transmembrane domain is present (fig. 2d). Obviously, such a protein loses its normal function as contact with the nerve cell is lost. It is intriguing, however, that truncated soluble forms of L1 still seem to be able to activate FGFRs and stimulate neurite outgrowth in vitro, although they miss the transmembrane and cytoplasmic domain [43]. This might suggest that L1 has essential functions not mediated through the FGFR signal transduction pathway. It is, however, also possible that the naturally occurring truncated L1 proteins in these patients are unstable and degraded quickly.
Genotype-Phenotype Correlation
Mutations in L1 cause a complex and variable spectrum of clinical abnormalities. However, no obvious relationship has been found between the disease phenotype and the type or position of the L1 mutation. Deletion of a large part of the cytoplasmatic domain, which would be likely to disrupt signaling or interactions with the cytoskeleton, has been observed in a family with a mild MASA phenotype [14, 32]. In contrast, missense mutations can lead to a very severe clinical picture [28, 31]. It is also unlikely that a good genotype-phenotype correlation exists, because of the large intrafamilial clinical variability with patients with HSAS, MASA, SP1 and ACC/DCC being present within the same family. Schrander-Stumpel et al. [pers. commun.] have reviewed the clinical characteristics of many families with HSAS, MASA, SP1 and ACC/DCC, and found that most families have a mixed phenotype with large intrafamilial variability. Most families have been referred to as HSAS, MASA, SP1 or ACC/DCC, depending on the clinical phenotype of the proband. In many families, however, affected relatives had different phenotypes.
L1 is not the only gene showing pleiotropic manifestations. More than 100 genes are known in which different mutations cause more than one clinical disorder. One of the most interesting examples is the RET protooncogene. RET is a transmembrane protein, with a cadherin-like extracellular domain, and a cysteine-rich extracellular region next to the transmembrane region. Although RET mutations cause four different clinical disorders, there is a good genotype-phenotype correlation. Inactivating mutations in several different domains of RET cause Hirschsprung’s disease, a developmental disorder of the enteric nervous system. Missense mutations of specific cysteine residues in the cysteine-rich region cause either multiple endocrine neoplasia type 2A and 2B, or medullary thyroid carcinoma through a dominant-negative oncogenic mechanism [45]. In contrast, the different L1 mutations do not cause a constant phenotype. The complicated signal transduction pathway, of which L1 is one of the many components, might be redundant and allow for variation in the expression of a loss-of-function mutation in L1 by the varying influence of other components. On the other hand, gain-of-function mutations might also have a variable clinical picture. Understanding the clinical variation encountered in CRASH syndrome might increase when we know more about the functioning of L1. The interactions of L1 with FGFR open another exciting field to be investigated. Recently, several mutations in the genes encoding FGFR 1 and 2 have been discovered, each of them associated with different craniosynostosis syndromes including Apert, Crouzon, Jackson-Weiss and Pfeiffer syndromes. Mutations in FGFR3, on the other hand, give rise to achondroplasia [46]. None of the latter syndromes show clinical overlap with CRASH syndrome, and clinical features such as craniosynostosis or dwarfism encountered in cases of an FGFR mutation have not been observed in families with an L1 mutation. This is not surprising, as until now L1 mutation analysis was only performed in families with CRASH features. It is possible that L1 is a candidate gene for Xq28-linked diseases with bone malformations. In this respect, it would be interesting to look for mutations in patients with oto-palato-digital syndrome, a bone disorder gene located in Xq28.
Differential Diagnosis
In view of the large clinical spectrum of the CRASH syndrome, the clinical diagnosis can be difficult, especially when hydrocephalus and adducted thumbs are absent. Nearly all patients have moderate to severe mental retardation, but it is still unclear how frequently L1 mutations will be found in nonspecific Xq28-linked mental retardation, but without hydrocephalus, spastic paraparesis or adducted thumbs [47]. It is possible, therefore, that patients with LI defects are not recognised as having CRASH syndrome. When hydrocephalus is present and the diagnosis CRASH is suspected, it is highly probable that an L1 mutation will be present. However, Strain et al. [48] have postulated the existence of a second locus for CRASH, approximately 10 cM proximal from the L1 gene. The linkage analysis upon which the authors based their findings, however, was remarkable, in that it showed an impressive number of intermarker recombinants over a short distance. Subsequently, a L1 mutation was identified in this family, excluding the existence of the hypothesized second locus [Brock, pers. commun.]. In our own experience of more than 50 HSAS or MASA families with multiple affected cases, we have not found evidence for linkage heterogeneity. Only one small German family was found in which a presumably healthy normal male had the same Xq28 haplotype as his brother and a cousin who were both affected with HSAS [7]. However, the normal male refused brain imaging or IQ testing, so that it could not be excluded that he was mildly affected. In some cases of the CRASH syndrome, the typical facial features of fragile X syndrome including elongated face and large everted cars are present [5].
X-linked spasticity and mental retardation (SP1) due to mutations in L1 have to be differentiated from SP2 [19,20]. This can be difficult when SP1 is not associated with other symptoms from the CRASH clinical spectrum (table 2). However, mental retardation is always moderate to severe in the CRASH syndrome and mild in SP2. The gene responsible for SP2 is PLP, located in Xq22. The PLP genes has two gene products, produced by alternative splicing: DM20, involved in oligodendrocyte maturation, and PLP itself, involved in myelin sheet compaction. Mutations affecting both gene products lead to Pelizaeus-Merzbacher disease, whereas mutations in PLP but spliced out in DM20 lead to SP2 [21].
Prenatal Diagnosis
In large families with HSAS, MASA, SP1 or ACC/DCC linked to the Xq28 region, DNA diagnosis based upon DNA analysis is possible as the Xq28 region contains many highly polymorphic markers. RFLP analysis can be performed using probe F814, which is a hybrid probe recognizing both a VNTR marker DXS52 and a two-allellic RFLP in F8C. DXS52 is approximately 1–2 cM proximal to L1, whereas F8C is 1–2 cM distal to L1. L1 itself is not very polymorphic as only one RFLP with a very low polymorphism information content has been found in L1 [49]. Nowadays, polymorphic microsatellites, amplified by PCR, are preferred, because this speeds up the analysis and requires a smaller amount of DNA, whereas microsatellites are much more informative than RFLP markers. In somes cases, a reliable diagnosis with flanking markers is not possible, as (1) the distal Xq28 markers are not always informative, (2) not enough DNA samples are available and (3) there is a de novo mutation. In such cases, it is necessary to identify the mutation itself to be able to perform prenatal diagnosis and carrier detection.
Conclusion
Mutations in the neural cell adhesion molecule L1, located in Xq28, can lead to a wide clinical spectrum of neurologic anomalies including corpus callosum hypoplasia, mental retardation, adducted thumbs, spastic paraparesis and hydrocephalus. We therefore propose to refer to this clinical spectrum as the CRASH syndrome.
References
Bickers DS, Adams RD: Hereditary stenosis of the aqueduct of Sylvius as a cause of congenital hydrocephalus. Brain 1949;72:246–262
Landrieu P, Ninane J, Ferriere G, Lyon G: Aqueductal stenosis in X-linked hydrocephalus: A secondary phenomenon? Dev Med Child Neurol 1979;21:637–652
Halliday J, Chow CW, Wallace D, Dansk DM: X-linked hydrocephalus: A survey of a 20 year period in Victoria, Australia. J Med Genet 1986;23:23–31
Fried K: X-linked mental retardation and/or hydrocephalus. Clin Genet 1972;3:258–263
Willems PJ, Brouwer OF, Dijkstra I, Wilmink J: X-linked hydrocephalus. Am J Med Genet 1987;27:921–928
Willems PJ, Dijkstra I, Van der Auwera BJ, Vits L, Coucke P, Raeymakers P, Van Broeckhoven C, Consalez GG, Freeman SB, Warren ST, Brouwer OF, Brunner HG, Renier WO, Van Elsen AF, Dumon JE: Assignment of X-linked hydrocephalus to Xq28 by linkage analysis. Genomics 1990;8:367–370
Willems PJ, Vits L, Raymakers P, Beuten J, Coucke C, Holden JJ, Van Broeckhoven C, Warren ST, Sagi M, Robinson D, Dennis N, Friedman KS, Magnay D, Lyonnet S, White BN, Wittwer BH, Aylsworth AS, Reicke S: Further localization of X-linked hydrocephalus in the chromosomal region Xq28. Am J Hum Genet 1992;51:307–315
Lyonnet S, Pelet A, Briard ML, Serville F, Le Marec B, Pfeiffer RA, Hors MC: Further linkage data at the X-linked hydrocephalus locus with seven polymorphic DNA markers of Xq28. Am J Med Genet 1991;49(suppl):A1959.
Bianchine JW, Lewis RC: The MASA syndrome: A new hereditary mental retardation syndrome. Clin Genet 1974;5:298–306
Gareis FJ, Mason MD; X-linked mental retardation associated with bilateral clasp thumb anomaly. Am J Med Genet 1984;17:333–338.
Yeatman GW: Mental retardationclasped thumb syndrome. Am J Med Genet 1984;17:339–344
Kenwrick S, Ionanescu V, Ionanescu G, Searby C, King A, Dubowitz M, Davies KE: Linkage studies of X-linked recessive spastic paraplegia using DNA probes. Hum Genet 1986;73:264–266
Straussberg R, Blatt I, Brand N, Kessler D, Goodman RM: X-linked mental retardation with bilateral clasped thumbs: Report of another affected family. Clin Genet 1991,40: 337–341.
Macias VR, Day DW, King TE, Wilson GN: Clasped thumb-mental retardation (MASA) syndrome: Confirmation of linkage to Xq28. Genetics 1992;43:408–414
Boyd E, Schwartz CE, Schroer RJ, May MM, Shapiro SD, Arena F, Lubs HA, Stevenson RE: Agenesis of the corpus callosum associated with MASA syndrome. Clin Dysmorphol 1993;2:332–341
Schrander-Stumpel C, Legius E, Fryns JP, Cassiman JJ: MASA syndrome: New clinical features and linkage analysis using DNA probes. J Med Genet 1990;27:688–692
Winter RM, Davies KE, Bell MV, Huson SM, Patterson MN: MASA syndrome: Further clinical delineation and chromosomal localisation. Hum Genet 1989;83:367–370
Harding AE: Classification of the hereditary ataxias and paraplegias. Lancet 1983;i:1151–1155
Keppen LD, Leppert MF, O’Connel PO, Nakamura D, Stauffer M, Lathrop M, Lalouel JM, White R: Etiological heterogeneity in X-linked spastic paraplegia. Am J Hum Genet 1987;41:933–943
Bonneau D, Rozet JM, Bulteau C, Berhier M, Metthey R, Munnich A, Le Merrer M: X linked spastic paraplegia (SPG2): Clinical heterogeneity at a single gene locus. J Med Genet 1993;30:381–384
Saugier-Veber P, Munnich A, Bonneau D, Rozet JM, Le Merrer M, Gil R, Boesplug-Tanguy O: X-linked spastic paraplegia and Pelizaeus-Merzbacher disease are allelic disorders at the proteolipid locus. Nat Genet 1994;6:257–262
Opitz JM, Richieri-da Costa A, Aase JM, Benke PJ: FG syndrome update 1988: Note of 5 new patients and bibliography. Am J Hum Genet 1988;30:309–328
Menkes JH, Philippart M, Clark DB: Hereditary partial agenesis of the corpus callosum. Arch Neurol 1964;11:198–208
Kang WM, Huang CC, Lin SJ: X-linked inheritance of dysgenesis of corpus callosum in a Chinese family. Am J Med Genet 1992;44:619–623
Kaplan P: X-linked recessive inheritance of agenesis of the corpus callosum. J Med Genet 1983;20:122–124
Rosenthal A, Jouet M, Kenwrick S: Aberrant splicing of neural cell adhesion molecule L1 mRNA in a family with X-linked hydrocephalus. Nat Genet 1992;2:107–112
Van Camp G, Vits L, Coucke P, Lyonnet S, Schrander-Stumpel C, Darby J, Holden J, Munnich A, Willems P: A duplication in the L1CAM gene associated with X-linked hydrocephalus. Nat Genet 1993;4:421–425
Jouet M, Rosenthal A, Macfarlane J, Kenwrick S, Donnai D: A missense mutation confirms the L1 defect in X-linked hydrocephalus. Nat Genet 1993;4:331.
Coucke P, Vits L, Van Camp G, Serville F, Lyonnet S, Kenwrick S, Rosenthal A, Wehnen M, Munnich A, Willems PJ: Identification of a 5′splice site mutation in intron 4 of the L1CAM gene in an X-linked hydrocephalus family. Hum Mol Genet 1994;3:671–673
Jouet M, Rosenthal A, Armstrong G, Macfarlane J, Stevenson R, Paterson J, Matzenberg A, Ionanescu V, Temple K, Kenwrick S: X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat Genet 1994;7:402–407
Fransen E, Schrander-Stumpel C, Vits L, Coucke P, Van Camp G, Willems PJ: X-linked hydrocephalus and MASA syndrome present in one family are due to a single missense mutation in exon 28 of the L1CAM gene. Hum Mol Genet 1994;3:2255–2256
Vits L, Van Camp G, Coucke P, Wilson G, Schrander-Stumpel C, Schwartz C, Willems P: MASA syndrome is due to mutations in the L1CAM gene. Nat Genet 1994;7:408–413
Moos M, Tacke R, Scherer H, Teplow D, Fruh K, Schachner M: Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domain similar to fibronectin. Nature 1988;334:701–702
Hlavin ML, Lemmon V: Molecular structure and functional testing of human L1CAM: an interspecies comparison. Genomics 1991;11:416–423
Lagenaur C, Lemmon V: An L1-like molecule, the 8D9 antigen, is a potent substrate for neurite extension. Proc Natl Acad Sci USA 1987;84:7753–7757
Wood PM, Schachner M, Bunge RP: Inhibition of Schwann cell myelination in vitro by antibody to the L1 adhesion molecule. J Neurosci 1990;10:3636–3645
Asou H, Miura M, Kobayashi M, Uyemura K, Itoh K: Cell adhesion molecule L1 guides cell migration in primary reaggregation culture of mouse cerebellar cells. Neurosci Lett 1992;144:221–224
Williams EJ, Doherty P, Turner G, Reid RA, Hemperly JJ, Walsh FS: Calcium influx into neurons can solely account for cell contact-dependent neurite outgrowth stimulated by transfected L1. J Cell Biol 1992;119:883–892
Persohn E, Schachner M: Immunoelectron microscopic localization of the neural cell adhesion molecules L1 and N-CAM during postnatal development of the mouse cerebellum. J Cell Biol 1987;105:569–576
Rathjen FG, Schachner M: Immunocytological and biochemical characterization of a new neuronal cell surface component (L1 antigen) which is involved in cell adhesion. EMBOJ 1984;3:1–10
Lemmon V, Farr KL, Lagenaur C: L1-mediated axon outgrowth occurs via a homophilic binding mechanism. Neuron 1989;2:1597–1603
Williams EJ, Furness J, Walsh FS, Doherty P: Activation of the FGF receptor underlies neurite outgrowth stimulated by L1, N-CAM and n-cadherin. Neuron 1994;13:583–594
Doherty P, Williams E, Walsh FS: A soluble chimeric form of L1 glycoprotein stimulates neurite outgrowth. Neuron 1995;14:57–66
Reid RA, Hemperly JJ: Variants of human L1-cell adhesion molecule arise through alternate splicing of RNA. J Mol Neurosci 1992,3:127–135.
Romeo G, McKusick VA: Phenotypic diversity, allelic series and modified genes. Nat Genet 1994,7: 451–453.
Mulvihill JJ: Craniofacial syndromes: No such thing as a single gene disease. Nature Genet 1995;9:101–103
Nordström AM, Penttinen M, von Koskull H: Linkage to Xq28 in a family with nonspecific X-linked mental retardation. Hum Genet 1992;90:263–266
Strain L, Gosden CM, Brock DJH, Bonthron DT: Genetic heterogeneity in X-linked hydrocephalus: Linkage to markers within Xq27.3. Am J Hum Genet 1994;54:236–243
Willems PJ, Vits L, De Boulle K: MspI RFLP in the L1CAM gene in Xq28. Nucleic Acids Res 1991;19:5448.
Acknowledgements
The research reviewed here was supported in part by the NFWO (National Fund for Scientific Research), the AFM (Association Française contre les Myopathies) and a concerted action from the University of Antwerp. E.F. holds a scholarship from the IWT (Institute for the Support of Research in Science and Technology). V.L. received support from the NEI (National Eye Institute) and NINDS (National Institute of Neurological Disorders and Stroke).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Fransen, E., Lemmon, V., Van Camp, G. et al. CRASH Syndrome: Clinical Spectrum of Corpus Callosum Hypoplasia, Retardation, Adducted Thumbs, Spastic Paraparesis and Hydrocephalus Due to Mutations in One Single Gene, L1. Eur J Hum Genet 3, 273–284 (1995). https://doi.org/10.1159/000472311
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472311
Key Words
This article is cited by
-
L1cam-mediated developmental processes of the nervous system are differentially regulated by proteolytic processing
Scientific Reports (2019)
-
A Fragment of Adhesion Molecule L1 Binds to Nuclear Receptors to Regulate Synaptic Plasticity and Motor Coordination
Molecular Neurobiology (2018)
-
Identification of novel mutations in L1CAM gene by a DHPLC-based assay
Genes & Genomics (2016)
-
Heterozygosity for the mutated X-chromosome-linked L1 cell adhesion molecule gene leads to increased numbers of neurons and enhanced metabolism in the forebrain of female carrier mice
Brain Structure and Function (2013)
-
Using C. elegans to Decipher the Cellular and Molecular Mechanisms Underlying Neurodevelopmental Disorders
Molecular Neurobiology (2013)