Abstract
Microsatellite analysis with 13 microsatellites spread over 18p was performed to determine the origin of the marker chromosome in 9 patients with additional metacentric marker chromosomes. Phenotypes and banding patterns suggested that the markers were isochromosomes 18p. Maternal origin was determined in all 8 cases where both parents were available for study. Six cases showed 3 alleles (one paternal, one maternal each in single and double dose) of informative markers located close to the telomere while markers close to the centromere on 18p were reduced to homozygosity (one paternal allele in single dosage and one maternal allele presumably in triple dosage). A similar result was obtained in the patient with no parents available for examination. The other 2 patients were uninformative for maternal hetero- versus homozygosity, but at some loci the maternal band was clearly stronger than the paternal one whereas the opposite was never observed. Trisomy 18 differs from trisomy 21, XXX and XXY of maternal origin through a preponderance of meiosis II versus meiosis I nondisjunction. Thus, the results of our study and the advanced mean maternal age at delivery of patients with additional i(18p) indicate that in most if not all cases the marker chromosome originates from maternal meiosis II nondisjunction immediately followed by isochromosome formation in one of the 2 maternal chromosomes 18. Possible explanations of these results include a maternally imprinted gene on 18q with a lethal effect if the paternal homologue is lost and a mechanism through which nondisjunction in some cases could be connected with isochromosome formation.
Similar content being viewed by others
Introduction
Supernumerary isochromosomes with one centromere or two centromeres in close proximity have so far been reported for 8p, 9p, 12p, 18p and Xq; in addition, there are instances of familial isochromosomes of short arms of acrocentrics containing only heterochromatic material. Supernumerary isodicentric chromosomes containing euchromatic material from the long arm were reported for 14(q11 and q12), 15(q12/13), 21(q11) and 22(q11) [1]. The most frequently observed autosomal isochromosomes appear to be i(12p), inv dup(15)(q12/13) and i(18p). The latter was first described in the prebanding era by Frøland et al. [2]. In this and subsequently reported cases, the additional marker was assumed to be an isochromosome 18p due to its size and (in later reports) banding pattern and due to the obvious consistency of the associated phenotype. Mattei et al. [3] were the first to demonstrate derivation from a chromosome 18 by in situ hybridization with the 18-specific probe D18S3. So far, clinical and/or cytogenetic data were reported on some 50 patients. The overwhelming majority of the cases occurred sporadically. Familial occurrences include 1 instance of maternal mosaicism [4] and 3 instances of a mother with trisomy 18p due to 18p deletion and isochromosome formation having given birth to a child with additional isochromosome 18p due to unbalanced transmission [5–7]. Several authors have observed an increased mean maternal age at delivery in patients with an additional isochromosome of the short arm of 18, i(18p) [8].
Here, we report the results of molecular studies on the origin and the mechanism of formation of an additional i(18p) chromosome in a single case and in 8 families with an affected child.
Patients and Methods
The Patients
All 9 patients disclosed clinical findings characteristic for tetrasomy 18p [1, 8]. Patients 1 and 6 have been reported previously [9, 10]. Data on age, sex, parental ages and clinical findings are presented in table 1.
Chromosome Examination and Fluorescence in situ Hybridization
Chromosomal analysis from preparations from lymphocyte cultures was performed in all patients and their parents by standard procedures (G bands using trypsin and Giemsa, GTG, and C bands using barium and Giemsa, CBG) at the level of 400 bands. A minimum of 50 metaphases were evaluated in each case. Fluorescence in situ hybridization (FISH) studies with a commercially available probe mapping to 18cen (D18Z1 from ONCOR) were performed on at least 20 metaphases in all 9 patients according to the protocols provided by the manufacturer.
Molecular Investigations
DNA was extracted from nucleated blood cells of all 9 cases and of the parents of cases 1–7 and 9 as previously described [11]. Micro-satellite loci as listed in table 2 were analyzed using the polymerase chain reaction (PCR). PCR amplification was done on an Perkin-Elmer thermocycler (32 cycles of 1 min at 94°C for denaturation, 1 min at 52–63°C for annealing, and 2 min at 72°C extension temperatures). 0.5–3 °l of reaction was mixed with an equal volume of urea loading buffer (42% urea, 0.1% xylene cyanol, 0.1% bromophe-nol blue and 0.1% of 0.5 M EDTA) and directly loaded onto a 0.4-mm-thick 6% polyacrylamide/50% urea gel. Bands were visualized by silver staining of the gels. A complete description of the allele frequencies and primers used to detect the loci is available from the Genome Database.
Results
Cytogenetic Findings
Chromosome examinations from lymphocyte cultures revealed a metacentric marker chromosome in addition to a normal karyotype in all investigated metaphases of cases 1–6 and 8–9. In case 7, the extra chromosome was present in one third (16/50) of lymphocyte metaphases. Banding patterns of the extra chromosomes confirmed that they were isochromosomes of the short arm of chromosome 18. All markers appeared monocentric in GTG-and CBG-banded preparations.
Metaphase chromosome FISH studies with the probe D18Z1 confirmed the origin of the additional chromosome from chromosome 18. In case 1–6, 8 and 9, all investigated metaphases from lymphocytes showed one additional signal.
Molecular Findings
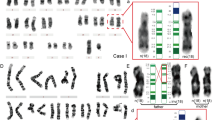
The molecular findings are listed on table 2. No parental DNA was available for study from patient 8. Six cases including case 8 showed 3 alleles at at least one locus mapping to distal 18p. For cases 1, 3, 4, 5, 7 and 9, 1 maternal allele of stronger intensity probably representing the isochromosome, 1 maternal allele of normal intensity and 1 paternal allele of normal intensity were observed (fig. 1a). In the remaining 2 cases (cases 2 and 6), 3 alleles were not observed for any marker, but when informative, the maternal allele always showed the stronger signal (fig. 1b). In addition, the results of examinations of 18q markers excluded maternal uniparental disomy 18q in all 7 cases examined, thus indirectly confirming the maternal origin of the marker chromosome. Thus, in the 8 families where both parents were investigated, the isochromosome of the proband originated from the mother. Markers on proximal 18p without exception showed reduction of maternal heterozygosity to homozygosity in the proband (fig. 1c). There was no marker constellation incompatible with paternity.
a Results for D18S59 in family 5. The patient has inherited the maternal allele a in single copy, the maternal allele c in double and the paternal allele d in single copy while he has not inherited the paternal allele b. b Results for D18S170 in family 9. The patient has inherited the maternal allele b, presumably in triple copy and the paternal allele a in single copy, c Results for D18S71 in family 9. The patient has inherited the maternal allele a, presumably in triple copy, and the paternal allele d in single copy while she has not inherited the maternal copy c and the paternal copy b; this indicates reduction of maternal heterozygosity to homozygosity at this locus close to the centromere of chromosome 18.
Discussion
Most authors have explained the formation of an isochromosome as a consequence of either misdivision at the centromere or a U-type exchange during meiosis or mitosis [12, 13]. A U-type exchange is more likely to result in a dicentric than in a monocentric isochromosome; however, all i(18p) chromosomes investigated by CBG banding appeared monocentric and thus are more likely to originate via misdivision at the centromere.
Theoretically, the isochromosome could be formed by the following mechanisms: (1) Trisomy 18 through nondisjunction first, centromeric misdivision second. In case of initial meiotic nondisjunction for chromosome 18, it should be possible to distinguish between errors occurring during meiosis I versus meiosis II through the study of molecular markers on proximal as compared to distal 18p. In case of meiosis I nondisjunction following recombination, heterozygosity of proximal and hetero- or homozygosity of distal markers should be observed while in case of meiosis II error following meiosis I recombination, the marker constellation should be reversed: homozygosity of informative markers at proximal and homo- or heterozygosity at distal 18p. The latter was demonstrated in all 7 patients in whom markers were informative for such a distinction. As no markers in the remaining 2 families were informative to show inheritance of both maternal alleles, these cases could also have resulted from an initial meiosis II segregational error. In addition, there was no case with 3 alleles at the loci very close to the centromere nor was there a case in which reduction of maternal heterozygosity to homozygosity in the proband was observed in a locus mapping distal to a locus with 3 alleles (table 2). If nondisjunction and misdivision at the centromere with loss of the long arm occurs postmeiotically, molecular analysis should reveal no more than 2 alleles, 1 in single copy and the other in triple copy, at any marker on 18p. This is excluded in 7 of the 9 cases of this report, the others being uninformative for this distinction.
(2) Misdivision first, nondisjunction second. This chronology of events appears unlikely as it would require two steps of abnormal cell division independent of each other yet closely following one another as monosomy for the entire 18q is most likely lethal for the cell. In the 7 cases with 3 alleles including 2 maternal ones at some distal loci, this mechanism of formation seems highly unlikely while in the 2 remaining cases it remains a remote possibility.
In summary, the results in the 7 informative families indicate that the isochromosome 18p is likely to commonly form through meiosis II nondisjunction immediately followed by meiotic or early postmeiotic mitotic misdivision at the centromere.
Furthermore, the results of molecular marker analysis in 8 families (proven origin of alleles in 6 and band intensity in the remaining 2) showed that all 8 isochromosomes are maternal in origin. Previous investigations of the origin of additional i(18p) chromosomes had been restricted to the demonstration of origin from a chromosome 18 through in situ hybridization [3, 14, 15] and had not included microsatellite analysis of the parental origin of the marker. However, in all families in whom the isochromosome 18p in 1 or several probands was the result of either a familial rearrangement [5–7] or of parental mosaicism [4], the origin was maternal. Indirect confirmation of the maternal origin of the marker in the 2 cases with no marker showing 3 alleles in the proband was obtained by exclusion of maternal uniparental disomy 18q through loss of the paternal 18q in these 2 and 5 other cases (table 1). Maternal meiotic nondisjunction is in agreement with the observed advanced mean maternal age both on the average of all cases (31.76 years, n = 47) and in the cases of this investigation in which a maternal origin of the i(18p) could be demonstrated (35.5 years, n = 8). This is significantly higher (p = 0.0043, t test) than the mean maternal age of 28 years in a Swiss control population [16].
At first it may seem surprising that all 6 informative isochromosomes should result from maternal meiosis II error. Maternal meiosis I is far more frequent than meiosisi II nondisjunction [for trisomy 21: Antonarakis et al., 17]. However, as recently demonstrated by Fisher et al. [18], the situation is different for trisomy 18. In their series of 51 cases of trisomy 18 due to maternal meiotic nondisjunction, they determined meiosis I error in only 16 and meiosis II error in 35. It seems that the special situation in trisomy 18 also accounts for those instances in whom one of the maternal chromosomes 18 is transformed to an isochromosome.
It is interesting to note that in none of the 8 cases in which both parents were available for study, the isochromosome was paternal and that in all cases there was biparental inheritance of the normal chromosomes No. 18. There are two possible explanations for this observation. One is that isochromosome formation occurs immediately after, and in connection (in an yet unknown way) with, (maternal) meiosis II nondisjunction and thus always affects one of the chromosomes involved in nondisjunction. The second is that there is a maternally imprinted gene on 18q which would not allow survival of the cell if the active paternal allele was lost. If the paternal chromosome 18 forms an isochromosome, with consecutive loss of the long arm, the cell would become maternally disomic for the entire 18q in addition to tetrasomy 18p. As deletions of 18q21.3→qter are not infrequent and, as evidenced from instances of unbalanced segregation of paternal balanced translocations, may be paternal in origin with no difference in clinical findings from the usual 18q phenotype [1, 8], and as interstitial deletions of 18q are extremely rare and their origin has not yet been studied, such an imprinted gene should map to the long arm proximal to 18q21.3. In contrast to many other human chromosomes including 13 and 21, uniparental disomy 18, either maternal or paternal, has so far not been demonstrated [19]. The results of examinations of 18q markers exclude maternal uniparental disomy 18q in 7 of the 9 patients and thus are compatible with this assumption.
The genetic map distance of the short arm of chromosome 18 is 54.1 cM for females [18]. This implies that one would expect on the average one recombination in the short arm of the maternal 18 in every meiosis. The observation of one recombination in 18p in all informative 6 (7 including the case without available parents and thus uncertainty about the origin of the marker) cases is thus in agreement with expectation and does not point towards the possibility of increased recombination causing meiotic nondisjunction and/or a possible mechanism by which recombination would be the basis for the nondisjunction combined with or immediately followed by centromeric misdivision.
In the available literature, we found only 1 case of an additional autosomal isochromosome in whom a similar study was performed. Fisher et al. [20] reported a case with mosaicism for an additional isochromosome 8p. The result of the molecular investigation was exactly the same as in the 6 fully investigated and informative i(18p) cases of this report: maternal origin of the marker, reduction of maternal heterozygosity to homozygosity in the proband for a marker closer to the centromere while a more distal marker showed one paternal and the two different maternal alleles, one in single and the other in double dosage. It will be interesting to examine other families with a proband mosaic for an additional isochromosome including i(12p) (the Pallister-Killian syndrome), i(8p) and i(9p) for the parent of origin and mechanism of formation in order to find out whether other mechanisms of origin for such markers exist.
References
Schinzel A: Human Cytogenetic Database. Oxford Medical Databases Series. Oxford, Oxford University Press, Electronic Publishing, 1994.
Frøland A, Holst G, Terslev E: Multiple anomalies associated with an extra small autosome. Cytogenetics 1963;2:99–106
Mattel MG, Philip N, Passage E, Moisan JP, Mandel JL, Mattel JF: DNA probe localization at 18p 113 band by in situ hybridization and identification of a small supernumerary chromosome. Hum Genet 1985;69:268–279
Abehovich D, Dagan J, Levy A, Steinberg A, Zlotogora J: Isochromosome 18p in a mother and her child. Am J Med Genet 1993;46:392–393
Uchida IA, Wang HC, Laxdal OE, Zaleski WA, Duncan BP: Partial trisomy-deficiency syndrome resulting from a reciprocal translocation in a large kindred. Cytogenetics 1964;3:81–96
Taylor KM, Wolfinger HL, Brown MG, Chadwick DL: Origin of a small metacentric chromosome: Familial and cytogenetic evidence. Clin Genet 1975;8:364–369
Takeda K, Okamura T, Hasegawa T: Sibs with tetrasomy 18p born to a mother with trisomy 18p. J Med Genet 1989;26:195–197
Schinzel A: A Catalogue of Unbalanced Chromosome Aberrations in Man. Berlin, Walter de Gruyter, 1984.
Schinzel A: Particular behavioural symptomatology in patients with rarer autosomal chromosome aberrations; in Schmid W, Nielsen J (eds): Human Behaviour and Genetics. Amsterdam, Elsevier, 1981, pp 195–210.
Rauch A, Pfeiffer RA, Trautmann U, Liehr U, Rott HD, Ulmer R: A study often small supernumerary (marker) chromosomes identified by fluorescence in situ hybridization (FISH). Clin Genet 1992;42:84–90
Miller SA. Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215.
de la Chapelle A: How do human isochromosomes arise? Cancer Genet Cytogenet 1982;5:173–179
Van Dyke DL, Babu VR, Weiss L: Letters to the editors: Parental age, and how extra isochromosomes (secondary trisomy) arise. Clin Genet 1987;32:76–80
Callen DF, Freemantie CJ, Ringenbergs ML, Baker E, Eyre HJ, Romain D, Haan EA: The isochromosome 18p syndrome: Confirmation of cytogenetic diagnosis in nine cases by in situ hybridization. Am J Hum Genet 1990;47:493–498
Blennow E, Nielsen KB: Molecular identification of a small supernumerary marker chromosome by in situ hybridization: Diagnosis of an isochromosome 18p with probe L1.84. Clin Genet 1991;39:429–433
Robinson WP, Lorda-Sanchez I, Malcolm S, Langlois S, Schuffenhauer S, Knoblauch H, Horsthemke B, Schinzel AA: Increased parental ages and uniparental disomy 15: A paternal age effect? Eur J Hum Genet 1993;1:280–286
Antonarakis SE: Down syndrome study group: Parental origin of the extra chromosome in trisomy-21 as indicated by analysis of DNA polymorphisms. N Engl J Med 1991;324:872–876
Fisher JM, Harvey JF, Morton NE, Jacobs PA: Trisomy 18: Studies of the parents and cell division of origin and the effect of aberrant recombination on nondisjunction. Am J Hum Genet 1995;56:669–675
Schinzel A, McKusick VA. Francomano C: Report of the Committee for Clinical Disorders and Chromosome Aberrations; in Cuticchia AJ (ed): Human Genetic Mapping 1994. A Compendium. Baltimore, The Johns Hopkins University Press, 1995, pp 1017–1071.
Fisher AM, Barber JCK, Crolla JA, James RS, Lestas AN, Jennings I, Dennis NR: Mosaic tetrasomy 8p: Molecular cytogenetic confirmation and measurement of glutathione reductase and tissue plasminogen activator levels. Am J Med Genet 1993;47:100–105
Acknowledgements
This project was supported by the Swiss National Foundation (grant No. 32-37798.93 to AS), The EMBO, Heidelberg (grant to DK) and the Stiftung für wissenschaftliche Forschung an der Universität Zürich (grant to AS). We express our gratitude to all the referring physicians, the patients and their parents.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kotzot, D., Bundscherer, G., Bernasconi, F. et al. Isochromosome 18p Results from Maternal Meiosis II Nondisjunction. Eur J Hum Genet 4, 168–174 (1996). https://doi.org/10.1159/000472191
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472191
Key Words
This article is cited by
-
Prenatal genetic diagnosis of tetrasomy 18p from maternal trisomy 18p: a case report
Molecular Cytogenetics (2022)
-
Pallister-Killian syndrome: clinical, cytogenetic and molecular findings in 15 cases
Molecular Cytogenetics (2018)
-
Adults with Chromosome 18 Abnormalities
Journal of Genetic Counseling (2015)
-
Tetrasomie 18p
Monatsschrift Kinderheilkunde (2008)
-
De novo isochromosome 18p in a female dysmorphic child
Journal of Applied Genetics (2006)