Abstract
BPES is a genetic disorder including blepharophimosis, ptosis of the eyelids, epicanthus inversus and telecanthus. Type I is associated with female infertility, whereas type II presents without other symptoms. Both types I and II occur sporadically or are inherited as an autosomal dominant trait. We present a molecular genetic and cytogenetic study in a large four-generation Belgian family with BPES type II. Karyotype analysis on high-resolution banded chromosomes yielded normal results. Fluorescence in situ hybridization (FISH) with cosmid probes spanning 3q22-q24 revealed normal hybridization patterns. Sixteen polymorphic CA repeats encompassing region 3q13-q25 were analysed. Linkage analysis in this large four-generation family provides conclusive evidence for the presence of a BPES gene in this region. Two-point lod scores greater than 3.0 between the disease and the following markers were seen: D3S1589 (4.67), D3S1292 (3.52), D3S1290 (3.59) and D3S1549 (3.65). By FISH, D3S1290, D3S1292 and D3S1549 were assigned to chromosome 3q23 using YACs positive for these markers.
Similar content being viewed by others
Introduction
Blepharophimosis syndrome (BPES, MIM 110100) is a rare eyelid malformation characterized by (1) a reduced size of the palpebral fissures (blepharophimosis), (2) falling of the eyelids (ptosis) and (3) a small skin fold arising from the lower lid and running inwards and upwards (epicanthus inversus) [1]. A lateral displacement of the inner canthi is also invariably present (telecanthus). BPES affects both sexes. Most cases are sporadic, but autosomal dominant inheritance has been observed in several families
BPES has been classified into two types: type I, with infertility in affected females, and type II, not associated with female infertility and transmitted by both sexes [2, 3]. Type I is completely penetrant, whereas type II has a penetrance of 96.5% [3, 4]. So far, it is not known whether female infertility is the result of a pleiotropic effect of the same gene or whether it is part of a contiguous gene syndrome.
Several sporadic BPES cases have been reported carrying an interstitial deletion of variable size involving 3q12-q24 [5–11]. Invariably, in these patients, BPES was associated with other anomalies such as mental retardation, a flat broad nasal bridge, muscular hypotonia, cryptorchidism, cardiac defects, a high arched palate, cleft of the soft palate, microcephaly and diaphragmatic hernia. This suggests the presence of a contiguous gene syndrome. It was recently shown that genetic heterogeneity might be present in BPES with some cases showing cytogenetic abnormalities at 3p25 [10].
In three unrelated male BPES children without other abnormalities, cytogenetic analysis revealed a balanced translocation involving 3q [12–14]. Classification into type I or II cannot be made in these cases.
One young girl with BPES and microcephaly as well as other minor anomalies was reported carrying a t(3;8)(q23;p21.1) translocation [15]. The authors suggested that this patient might have a microdeletion.
Linkage analysis with 3q markers was reported in two small BPES type II families with autosomal dominant inheritance [16]. Under the assumption of complete penetrance, a lod score of 3.23 was obtained by multipoint analysis, suggesting that a gene for BPES is located on chromosome 3q. However, an interval of approximately 50 cM was supported with 100:1 odds by the multipoint analysis. Recently, linkage analysis in a large French pedigree provided evidence for the localisation of a BPES gene in the 18-cM interval between D3S1292 and D3S1555 [17].
In this communication, we present our cytogenetic, linkage and FISH data obtained in a large Belgian BPES type II family.
Materials and Methods
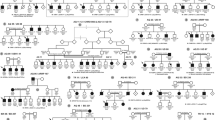
Careful clinical examination including measurements of the inner canthal distance was carried out in a core group of family members, including six affecteds, resulting in the diagnosis BPES type II [18]. Other family members were diagnosed on the basis of extensively available early photographs. Our patients have the classical eye abnormalities, but no other abnormalities besides a dry scaling skin. All affected persons had an affected parent and reduced penetrance was not obvious. High-resolution banding analysis was performed on metaphase spreads prepared from short-term phytohaemagglutininstimulated lymphocyte cultures by standard methods [19].
To detect submicroscopic deletions, cosmids cC13–438, cC13–493, CC13–596 and cC13–608 located in 3q22–q24 [20] were analysed by FISH as described elsewhere [21]. DNA was extracted using standard phenol-chloroform methods [22].
Segregation of sixteen microsatellite markers was studied in the family: D3S1558, D3S1303, D3S1267, D3S1589, D3S1292, D3S1290, D3S1576, D3S1549, D3S1593, D3S196, D3S1306, D3S1555, D3S1308, D3S1299, D3S1279 and D3S1275 [23]. The microsatellites were selected for their high heterozygosity value [23] and for their localization in the region 3q13–3q25 [24]. The CA strand primers were fluorescein labelled at the 5′ end. CA repeat fragments were amplified essentially as described in Gyapay et al. [23] in 25-µl reaction mixtures containing 100 ng DNA, 1 U Taq polymerase (BRL), 20 mM Tris-HCl (pH 8.8), 1.5 mM MgCl2 15 mM (NH4)22SO4, 0.1 % Triton X-100, 0.01 % gelatin, 200 mM each dNTP, 1 mM of each primer. For every marker, DNA amplified from individual 134702 of the CEPH family 1347 served as a size marker, allowing accurate genotyping of the patients [23]. All PCR fragments were electrophoresed on an ALF™ Automated DNA sequencer (Pharmacia) on a 6% LongRanger™-(AT Biochem) 8 M urea denaturing gel. In every lane, size markers (63 and 334 bp) were loaded to correct for minor bandshifts during gel electrophoresis. Genotyping for all markers was carried out using the Fragment Manager™ 1.1 software.
Lod scores were calculated using LINKAGE 5.1 [25], assuming a gene frequency of 0.0001 and a disease penetrance of 0.965. Allele frequencies for the different markers were taken from the Genome Data Base. Multipoint linkage analysis was performed using the Fast-link programmes [26, 27].
To determine the cytogenetic localization of the BPES type II region, several YACs positive for closely linked markers were identified from the Généthon database. Y730D4 (D3S1290 and D3S1587), Y819B5 (D3S1292), Y648G2 and Y710G8 (both D3S1549) were mapped by FISH on R-banded chromosomes. Probe labelling, slide preparation, pretreatment and FISH were performed as described elsewhere [21]. Images were recorded with the ISIS digital imaging system (MetaSystems).
Results
Analysis of high-resolution-banded chromosomes in affected and non-affected relatives was normal. No deletions were found by FISH with the cosmids cC13–438, CC13–493, CC13–596 and cC13–608 (data not shown).
For all markers tested, several affected persons were heterozygous, implying that in our family, BPES type II is not caused by a large deletion at these sequence-tagged sites.
The 2-point lod scores for the most informative markers are shown in table 1. With four adjacent markers, a 2-point lod score higher than 3 was obtained at θ = 0.00: D3S1589 (4.67), D3S1292 (3.52), D3S1290 (3.59) and D3S1549 (3.65).
Under the assumption of complete penetrance, the positive values of our 2-point lod scores rise further. Haplotype analysis revealed two key crossovers, allowing a definition of the interval for BPES type II by meiotic breakpoints to a 28-cM interval between D3S1593 and D3S1558. Multipoint analysis under the assumption of full penetrance showed a platform between D3S1593 and D3S1558 (fig. 1).
To determine the cytogenetic localization of the BPES type II region, four YACs positive for closely linked markers were selected from the CEPH YAC library using the Généthon Quickmap program. Y730D4 (D3S1290 and D3S1587), Y819B5 (D3S1292), Y648G2 and Y710G8 (both D3S1549) were mapped by FISH on R-banded chromosomes. Specific hybridization signals were observed on 3q23 for each of the four YACs (fig. 2). YACs 648G2 and 710G8 were chimeric: besides the hybridization to 3q23, a weak signal was seen at 18q for Y648G2 and at 3q, 8q, 14q and 1q for Y710G8.
Discussion
The patients in this family present with BPES type II, without other associated findings besides a dry scaling skin. As no cytogenetic abnormalities were observed and FISH analysis in the region 3q22-3q24 did not reveal a deletion, BPES type II is most probably caused by a single gene defect in this family.
Linkage analysis allowed the conclusion that a gene for BPES tpe II is located in this region. This is the first linkage report on an extended family resulting in a 2-point lod score of 4.67 between BPES type II and the marker D3S1589, under the assumption of reduced penetrance. We analysed our data under this most conservative and stringent assumption, because BPES type II families have been described where reduced penetrance was evident [3, 4]. Previous linkage studies were performed under the assumption of complete penetrance [16, 17], as all affected patients had an affected parent. In our opinion, however, this argument only means that reduced penetrance is not obvious.
Combining the results of the three studies [16, 17, present paper], under the assumption of full penetrance, we can refine the support interval for the BPES gene by linkage with odds of 100:1, to a 14-cM interval between D3S1292 and D3S1593. So far, linkage data in familial BPES type II, with no detectable cytogenetic aberration, reveal no genetic heterogeneity.
At present, no linkage studies have been reported in families with BPES type I. Although BPES type I is more prevalent than type II, it may be more difficult to encounter a family suitable for analysis, because of the female infertility.
The correlation between the cytogenetic and the genetic linkage map is not yet well established. By FISH, four YACs containing closely linked markers (D3S1292, D3S1290 and D3S1549) were precisely mapped on chromosome 3q23 and the localization of the markers was therefore refined compared to Smith et al. [24].
Recently, the translocation breakpoint in the patient described by Fukushima et al. [14] was localized between D3S1615 and D3S1316 [28]. Both markers lie 1 cM from the marker D3S1549, yielding a 3.65 2-point lod score in our family, and mapped to 3q23. We presume that in our family also, the defect might reside near the translocation breakpoint and we are currently investigating this region. The gene encoding the cellular retinol-binding protein-1 (RBP1), mapped to this region, represents a potential candidate for the BPES gene, as retinoids are required for a wide variety of biological processes. RBP1 is also considered to be a candidate gene for Hailey-Hailey, an autosomal dominant skin disease mapped to the same region as BPES type II [29]. The dry scaling skin in our BPES patients might be a minor expression of Hailey-Hailey. Further studies are needed to determine the potential involvement of RBP1 in both disorders.
References
Oley C, Baraitser M: Blepharophimosis, ptosis, epicanthus inversus syndrome (BPES syndrome). J Med Genet 1988;25:47–51
Townes P, Muechler E: Blepharophimosis, ptosis, epicanthus inversus and primary amenorrhea: A dominant trait. Arch Ophthalmol 1979;97:1664–1666
Zlotogora J, Sagi M, Cohen T: The blepharophimosis, ptosis and epicanthus inversus syndrome: Delineation of two types. Am J Hum Genet 1983;35:1020–1027
Temple I, Baraister M: Pitfalls in counselling of the blepharophimosis, ptosis, epicanthus inversus syndrome (BPES). J Med Genet 1989;26:517–519
Alvarado M, Bocian M, Walker A: Interstitial deletion of the long arm of chromosome 3: Case report, review and definition of a phenotype. Am J Med Genet 1987;27:781–786
Fryns JP, Stromme P, van den Berghe H: Further evidence for the localisation of the blepharophimosis syndrome (BPES) at 3q22.3–q23. Clin Genet 1993;44:149–151
Fujita H, Meng J, Kawamura M, Tozuka N, Ishii F, Tanaka N: Boy with a chromosome del(3)(q12q23) and blepharophimosis syndrome. Am J Med Genet 1992;44:434–436
Ishikiriyama S, Goto M: Blepharophimosis sequence (BPES) and microcephaly in a girl with del(3)(q22.2q23): A putative gene responsible for microcephaly close to the BPES gene? Am J Med Genet 1993;47:487–489
Jewett T, Rao P, Weaver R, Stewart W, Thomas I, Pettenati M: Blepharophimosis, ptosis and epicanthus inversus syndrome (BPES) associated with interstitial deletion of band 3q22: Review and gene assignment to the interface of band 3q22.3 and 3q23. Am J Med Genet 1993;47:1147–1150
Warburg M, Bugge M, Brondum-Nielsen K: Cytogenetic findings indicate heterogeneity in patients with blepharophimosis, epicanthus and developmental delay. J Med Genet 1995;32:19–24
Wolstenholme J, Brown J, Masters K, Wright C, English C: Blepharophimosis sequence and diaphragmatic hernia associated with interstitial deletion of chromosome 3 (46, XY, del(3)(q21q23)). J Med Genet 1994;31:647–648.
Boccone L, Meloni A, Falchi A, Usai V, Cao A: Blepharophimosis, ptosis, epicanthus inversus syndrome, a new case associated with de novo balanced autosomal translocation [46,XY,t(3;7)(q23;q32)]. Am J Med Genet 1994;51:258–259
De Die-Smulders C, Engelen J, Donk J, Fryns J: Further evidence for the location of the BPES gene at 3q2. J Med Genet 1991;28:725.
Fukushima Y, Wakui T, Nishida T, Ueoka Y: Blepharophimosis sequence and de novo balanced autosomal translocation [46,XY, t(3;4)(q23;p15.2)]: Possible assignment of the trait to 3q23. Am J Med Genet 1991;40:485–487.
Cabral de Almeida J, Llerena J, Neto J, Jung M, Martins R: Another example favouring the location of BPES at 3q2. J Med Genet 1993;30:86–88
Small K, Stalvey M, Fisher L, Mullen L, Dickel C, Beadles K, Reimer R, Lessner A, Lewis K, Pericak-Vance M: Blepharophimosis syndrome is linked to chromosome 3q. Hum Mol Genet 1995;4:443–448
Amati P, Chomel JC, Nivelon-Chevalier A, Gilgenkrantz S, Kitzis A, Kaplan J, Bonneau D: A gene for blepharophimosis-ptosis-epicanthus inversus syndrome maps to chromosome 3q23. Hum Genet 1995;96:213–215
Kohn R, Romano P: Blepharoptosis, blepharophimosis, epicanthus inversus and telecanthus — A syndrome with no name. Am J Ophthalmol 1971;72:625–632
Yunis JJ: High resolution human chromosomes. Science 1976;191:1268–1270
Yamakawa K, Morita R, Takahashi E, Hori T, Lathrop M, Nakamura Y: A genetic linkage map of 41 restriction fragment length polymorphism markers for chromosome 3. Genomics 1991;11:565–572
Van Roy N, Laureys G, Versteeg R, Opdenakker G, Speleman F: High resolution fluorescence mapping of 48 markers to the short arm of human chromosome 1. Genomics 1993;18:71–78
Sambrook J, Fritsch E, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, Cold Spring Harbor Laboratory, 1989.
Gyapay G, Morisette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J: The 1993–94 Généthon human genetic linkage map. Nat Genet 1994;7:246–339
Smith D, Glover W, Gemmill R, Drabkin H, O’Connell P, Naylor S: Report of the fifth international workshop on human chromosome 3 mapping 1994. Cytogenet Cell Genet 1995;68:126–146
Lathrop G, Lalouel J: Easy calculation of lod scores and genetic risks on small computers. Am J Hum Genet 1984;36:460–465
Lathrop G, White R: Construction of human genetic linkage maps: Likelihood calculations for multilocus analysis. Genet Epidemiol 1986;3:39–52
Cottingham R, Idury R, Schaffer A: Faster sequential genetic linkage computations. Am J Hum Genet 1993;53:252–263
Lawson C, Toomes C, Fryer A, Carette M, Taylor G, Fukushima Y, Dixon M: Definition of the blepharophimosis, ptosis, epicanthus inversus syndrome critical region at chromosome 3q23 based on the analysis of chromosomal anomalies. Hum Mol Genet 1995;4:963–967
Ikeda S, Welsh E, Peluso A, Leyden W, Duvic M, Woodley D, Epstein E: Localisation of the gene whose mutations underlie Hailey-Hailey disease to chromosome 3q. Hum Mol Genet 1994;3:1147–1150
Acknowledgements
The authors thank Dr. K. Hashimoto for providing chromosome 3 cosmids and the Fondation Jean Dausset-CEPH for chromosome 3 YACs. This work was partially supported by the National Fund for Scientific Research, Belgium.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Messiaen, L., Leroy, B.P., De Bie, S. et al. Refined Genetic and Physical Mapping of BPES Type II. Eur J Hum Genet 4, 34–38 (1996). https://doi.org/10.1159/000472167
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472167