Abstract
Aim:
Daidzein (4′,7-dihydroxyisoflavone) is an isoflavone exiting in many herbs that has shown anti-inflammation activity. The aim of this study was to investigate the mechanism underlying its anti-inflammatory action in murine lung epithelial cells.
Methods:
C57BL/6 mice were intranasally exposed to TNF-α to induce lung inflammation. The mice were injected with daidzein (400 mg/kg, ip) before TNF-α challenge, and sacrificed 12 h after TNF-α challenge, and lung tissues were collected for analyisis. In in vitro studies, murine MLE-12 epithelial cells were treated with TNF-α (20 ng/mL). The expression of pro-inflammatory chemokine Cxcl2 mRNA and NF-κB transcriptional activity were examined using real-time PCR and a dual reporter assay. Protein poly-adenosine diphosphate-ribosylation (PARylation) was detecyed using Western blotting and immunoprecipitation assays.
Results:
Pretreatment of the mice with daidzein markedly attenuated TNF-α-induced lung inflammation, and inhibited Cxcl2 expression in lung tissues. Furthermore, daidzein (10 μmol/L) prevented TNF-α-induced increases in Cxcl2 expression and activity and NF-κB transcriptional activity, and markedly inhibited TNF-α-induced protein PARylation in MLE-12 cells in vitro. In MLE-12 cells co-transfected with the PARP-1 expression plasmid and NF-κB-luc (or Cxcl2-luc) reporter plasmid, TNF-α markedly increased NF-κB (or Cxcl2) activation, which were significantly attenuated in the presence of daidzein (or the protein PARylation inhibitor PJ 34). PARP-1 activity assay showed that daidzein (10 μmol/L) reduced the activity of PARP-1 by ∼75%.
Conclusion:
The anti-inflammatory action of daidzein in murine lung epithelial cells seems to be mediated via a direct interaction with PARP-1, which inhibits RelA/p65 protein PARylation required for the transcriptional modulation of pro-inflammatory chemokines such as Cxcl2.
Similar content being viewed by others
Introduction
Daidzein (4′,7-dihydroxyisoflavone) is a small molecule that belongs to the group of phytoisoflavones and has been widely used in the treatment of various diseases such as infectious or non-infectious inflammation1,2,3. Soybeans, which are most widely used in Asian countries, are rich sources of biologically active isoflavones such as daidzein. Recently emerging studies have been conducted to fully exploit the anti-inflammatory properties of this molecule. It was reported that daidzein at a concentration of 10 μmol/L increased IL-2, IL-4, and IFN-γ production in phytohemagglutinin-stimulated peripheral blood mononuclear cells and reduced IL-10 and IFN-γ levels at higher concentrations (>50 μmol/L) in a senescence-accelerated-prone mouse model4. The effect of daidzein on human dendritic cell (DC) activation via lipopolysaccharide (LPS) stimulation and the subsequent DC-mediated effecter cell function both in vitro and in a mouse model of upper airway inflammation has been investigated. Daidzein treatment suppressed the expression of DC maturation markers (CD83, CD80, CD86) and MHC class I molecules, as well as the mucosal immune response, in ovalbumin-sensitized mice5. It was also reported that daidzein significantly inhibited the production of NO and IL-6, as well as their mRNA expression, in LPS-treated RAW264.7 cells6. However, the exact molecular mechanisms for the immune inhibition of daidzein remain elusive, which has hampered its further application in disease treatment.
Poly-adenosine diphosphate-ribosylation (PARylation) is a process in which NAD+ is used as a substrate, resulting in the formation of poly-adenosine diphosphate ribose (PAR), and the polymers are covalently attached to the receptor proteins via a family of poly-ADP-ribose polymerases (PARP)7,8. PARP-1 is the most characterized member of this family and mediates 85% of its activity. The importance of PAR synthesis has been established in many cellular processes, including DNA repair, chromatin replication, transcriptional regulation, and cell death8,9,10,11. A growing list of evidence from animal models has demonstrated involvement of the enzymatic activation of PARP-1 in the progression of inflammatory disorders. The main causes for the roles of PARP-1 in inflammatory pathogenesis are activated PARP-1 regulating pro-inflammatory gene expression and excessively activated PARP-1 overconsuming the intracellular ATP stores12. Our previous studies and those of others have shown that PARP-1 activation contributed to the enhanced expression of proinflammatory cytokines, eg, IL-1β and TNF-α13, inflammation-relevant adhesion molecules, such as ICAM-1, P-selectin, and E-selectin14,15, and the activation of enzymes was involved in the deleterious effects of inflammation, eg, iNOS, COX-2 and NADPH oxidase15,16,17,18, all of which promote the pathogenesis of inflammatory diseases.
Isoflavones such as daidzein exert their anti-inflammatory properties in various disease models19,20. We hypothesize that the anti-inflammatory effect of daidzein is mediated by impairing the activity of PARP, which would thereby decrease the expression of inflammatory genes. To examine this possibility, we utilized TNF-α-challenged mice, murine airway epithelial MLE-12 cells and a variety of molecular approaches.
Materials and methods
Reagents and antibodies
Daidzein (572667) and PJ34 (P4365, PARP-1 inhibitor) were purchased from Sigma-Aldrich. TNF-α was purchased from PeproTech Inc (Rocky Hill, NJ, USA). A monoclonal antibody against polyADP-ribose (PAR) (ALX-804-220) was purchased from Alexis (San Diego, CA, USA), and monoclonal antibodies against PARP-1 (B-10, sc-74470) and NF-κB subunit p65 (RelA) (F-6, sc-8008) were purchased from Santa Cruz Biotech (Santa Cruz, CA, USA).
Mice and TNF-α exposure
Eight-week-old male C57BL/6 mice (∼20 g) were maintained in a specific pathogen-free facility in the Medical School of Jilin University (Changchun, China) and allowed free access to standard rodent food and water. The mice were exposed to a single dose of challenge by intranasally introducing 50 μL of TNF-α (200 ng/mL in saline), as described in our previous study21. One hour before TNF-α challenge, the mice were pretreated with or without daidzein (400 mg/kg)22 or PJ34 (10 mg/kg)23 by intraperitoneal administration. All administration and surgical procedures were approved by the Experimental Animal Committee of Jilin University.
Lung histology
The mice were sacrificed 12 h after TNF-α challenge (n=3), and lung tissues were collected and fixed in 10% buffered formalin. Lung sections (5-μm thick) were prepared, and histological examination of inflammatory cell filtration was performed using hematoxylin and eosin (H&E) staining. Images were captured at 20× magnification using a Nikon 80i microscope (Nikon, Tokyo, Japan).
Cell culture and stimulation
The murine MLE-12 epithelial cell line was cultured in DMEM/F12 (Gibco, Grand Island, NY, USA) containing 2% fetal calf serum. The cells were incubated in the presence of TNF-α (20 ng/mL) for 1 or 6 h. For inhibitory experiments, the cells were incubated with TNF-α in the presence of a specific inhibitor of PARP, PJ34 (2.5 μmol/L), or daidzein (10 μmol/L).
Constructs
The mouse Cxcl2 promoter (-571 to +81) was cloned from the MLE-12 genome and inserted into the reporter vector pGL4.20 (Promega, Madison, WI, USA) using the restriction enzyme sites Kpn I and Bgl II to generate the construct Cxcl2-Luc. A NF-κB reporter plasmid containing 5× NF-κB binding sites (TGGGGACTTTCCGC)5 was kindly provided by Dr Istvan BOLDOGH (University of Texas Medical Branch). The plasmid pRL-SV40, which encoded Renilla luciferase driven by the SV40 promoter (Promega), was used as an internal control. A PARP-1 expression plasmid within a pcDNA background was a generous gift of Dr Patrick A ZWEIDLER-MCKAY (The University of Texas MD Anderson Cancer Center).
Immunoblotting
MLE-12 cells (2×106 per sample) were cultured and stimulated with TNF-α in the presence or absence of PJ34 or daidzein and lysed in lysis buffer [50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Nonidet P-40, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L glycerophosphate, 1 mmol/L Na3VO4, 1 mmol/L NaF, and 20 μg/mL aprotin/leupeptin/PMSF]. Twenty micrograms of protein from each sample was resolved using SDS-PAGE. After the proteins were transferred to nitrocellulose membranes, the membranes were washed with TBST [20 mmol/L Tris base, 500 mmol/L NaCl, 0.05% Tween-20 (pH 7.5)], blocked with 5% non-fat dry milk and then incubated with primary antibody against PAR and horseradish peroxidase-conjugated secondary antibody for 1 h each. Signals were detected using the ECL plus chemiluminescent detection system (Amersham).
Immunoprecipitation
MLE-12 cells (1×107 per sample) were cultured, stimulated as described above, and lysed in lysis buffer. The lysates were centrifuged at 4 °C and 13 000×g for 30 min, and the supernatants were incubated with 30 μL of protein G-Sepharose (Millipore Corporation Billerica, MA, USA) at 4 °C for 2 h. The pre-cleared supernatants were incubated with the antibodies against PARP-1 or RelA/p65 for 12 h and then with protein G-Sepharose for 2 h with continuous rotation. Immunoprecipitates were then washed with lysis buffer and resolved by SDS-PAGE, and the proteins were transferred to nitrocellulose membranes. The nitrocellulose membranes were washed with TBST, blocked with 5% non-fat dry milk and then incubated with antibodies against PARP-1, p65, or PAR, and horseradish peroxidase-conjugated secondary antibody for 1 h each. The signals were detected using the ECL plus chemiluminescent detection system (GE Life Sciences, Bucking Hampshire, UK).
Reverse transcription and real-time PCR
Total RNA from mouse lung or MLE-12 cells after various treatments was extracted using TRIzol reagent (Invitrogen), and 2 μg of the purified RNA from each sample was transcribed into cDNA. The cDNA was used as template, and real-time PCR was performed using an ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). The following pairs of primers were used for the expression of Cxcl2 and GAPDH: Cxcl2 forward: 5′-CTCCTTTCCAGGTCAGTTAGC-3′, reverse: 5′-CAGAAGTCATAGCCACTCTCAA-3′; GAPDH forward: 5′-CTCATGACCACAGTCCATGC-3′; reverse: 5′-CACATTGGGGGTAGGAACAC-3′.
Transient transfection and luciferase reporter assay
MLE-12 cells were transiently transfected with the reporter or expression plasmids using Lipofectamine 2000 (Invitrogen) following the manufacturer's instruction. The cells were seeded in 24-well plates, incubated in growth medium without antibiotics overnight and then transfected with the Cxcl2-luc or pNF-κB-luc (800 ng) and pRL-SV40 (100 ng) plasmids. After 6 h, the cells were replaced with complete medium to promote recovery. Twelve hours later, the transfected MLE-12 cells were stimulated with or without TNF-α (in the presence or absence of PJ34 or daidzein) for 6 h and then lysed. Firefly and Renilla luciferase activity was measured using the dual luciferase assay system (Promega) and a luminometer. Transcriptional activity was represented by the luciferase activity of firefly normalized to that of Renilla. For co-transfection experiments, expression plasmids encoding human PARP-1 or pcDNA3 (negative control) were co-transfected with the reporter plasmids, and the transcriptional activity was measured as described above.
In vitro PARP-1 activity detection
A Trevigen PARP-1 activity detection kit was utilized to examine the inhibition of daidzein on PARP-1 activity according to the protocols provided by the manufacturer. To quantify the inhibitory effect, luminescence representing the activity of PARP-1 was read using a Centro XS3 LB 960 (Berthold Technologies).
Statistical analysis
All experiments were performed at least thrice for each determination. Data were expressed as the mean±SD and analyzed using one-way ANOVA. The level of significance was accepted at P<0.01.
Results
Daidzein attenuates TNF-α-induced inflammation in mice lungs
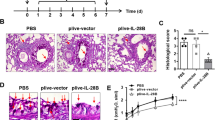
To examine the anti-inflammatory role of daidzein, we intranasally challenged mice with TNF-α with or without pre-administration of daidzein intraperitoneally, as described in Materials and methods. The upper one third of the lungs was harvested 1 h later, RNA was extracted, and cDNA was used as a template to carry out real-time PCR. TNF-α-challenge increased the mRNA levels (more than 200-fold) of Cxcl2 [homolog of human growth-regulated protein beta (Gro-β)], which is a potent attractant of neutrophils (Figure 1A). Daidzein administration significantly decreased the level of Cxcl2 mRNA (by ∼78%±9.02%). Accordingly, 12-h TNF-α exposure caused marked inflammatory cell infiltration in the lungs of mice, as shown by H&E staining (Figure 1Bb), and this effect was significantly attenuated by daidzein administration (Figure 1Bc). In selected experiments, mice were pre-treated intraperitoneally with the protein PARylation inhibitor PJ34, which inhibited the increase in the level of Cxcl2 mRNA to nearly basal levels (Figure 1A) and blocked the recruitment of inflammatory cells to mouse lungs (Figure 1Bd).
Daidzein attenuates lung inflammation induced by TNF-α. Mice were challenged intranasally with TNF-α (10 ng in 50 μL of saline) with or without intra-peritoneal administration of daidzein (400 mg/kg) or PJ34 (10 mg/kg) 1 h before TNF-α challenge. (A) The mice were sacrificed 1 h later, the upper one third of the lungs was harvested, and RNA was extracted and reverse transcribed. Real-time PCR was performed using pooled cDNA as a template to analyze the Cxcl2 mRNA level, n=5 (cP<0.01 compared with normal mice; fP<0.01 compared with TNF-α-challenged mice). (B) The mice were sacrificed 12 h later, and the lung sections were subjected to H&E staining (bar, 50 μm).
Daidzein decreased mRNA levels of the chemokine Cxcl2 induced by TNF-α
Because bronchoalveolar epithelial cells are critical defense barrier against exogenous pathogens and are implicated in the initiation of inflammation in lungs by TNF-α and under various pulmonary disease conditions24,25,26, we utilized murine MLE-12 epithelial cells to obtain insight into the molecular mechanism by which daidzein inhibits inflammation. MLE-12 cells were transfected with the Cxcl2-luc reporter plasmid and then exposed to TNF-α (20 ng/mL) in the presence or absence of daidzein or PJ34. TNF-α exposure induced a >5-fold increase in transcription from the Cxcl2 promoter, and this effect was significantly inhibited by daidzein (10 μmol/L) and PJ34 (2.5 μmol/L) (40% and 30%, respectively) (Figure 2A). The proximal region of the mouse Cxcl2 promoter contains two NF-κB binding sites, so, in support, we further showed a >10-fold increase in Luc activity that was driven by a promoter containing the NF-κB-binding consensus. Daidzein treatment of cells decreased TNF-α-induced transcription by 70% (Figure 2B), and PJ34 treatment nearly inhibited TNF-α-induced activation from the NF-κB binding site-containing promoter, which was in line with our previous result27. Likewise, the mRNA level of the chemokine Cxcl2 was also upregulated more than 40-fold upon TNF-α simulation, and this effect was diminished by treatment with daidzein or PJ34 to approximately baseline levels (Figure 2C). Moreover, a dose-dependent response of the inhibitory effect was observed when daidzein was administered at concentration ranges of 0.5, 1, 2, 5, and 10 μmol/L (Figure 2D).
Daidzein decreases TNF-α -dependent Cxcl2 expression. MLE-12 cells were transiently transfected with the reporter plasmids (A) Cxcl2-luc or (B) NF-κB-luc and Renilla-luc, as described in Materials and methods. The cells were cultured and stimulated with TNF-α in the presence or absence of daidzein (10 μmol/L) or PJ34 (2.5 μmol/L) for 6 h. Cxcl2-driven Luc expression and NF-κB transcriptional activation was measured by normalizing the firefly luciferase activity to that of renilla luciferase. The expression in untreated cells was considered 1. (C) MLE-12 cells were exposed or not exposed to TNF-α (in the presence or absence of daidzein or PJ34) for 1 h, RNA was extracted, and real-time PCR was performed to analyze Cxcl2 mRNA levels. (D) MLE-12 cells were exposed to TNF-α for 1 h in the presence of different concentrations of daidzein, as indicated, RNA was extracted, and real-time PCR was performed to analyze Cxcl2 mRNA levels. Data (mean±SD) are representative of three independent experiments (cP<0.01 compared with normal cells; fP<0.01 compared with TNF-α-stimulated cells).
Daidzein blocks the transcriptional activation of pro-inflammatory genes via inhibiting PARP-1 activity
Because protein PARylation is known to contribute to the pathogenesis of inflammatory disorders12, we assessed the capacity of daidzein to inhibit protein PARylation caused by TNF-α exposure in murine MLE-12 epithelial cells. In TNF-α-treated cells, protein PARylation was notably increased beginning at 1 h, and it reached a maximum at 6 h post exposure (Figure 3A), which was in line with our previous result obtained from the study of macrophages27. Daidzein exhibited a substantial inhibitory effect on PARP activity (Figure 3B) at the 6-h time point.
Daidzein blocks pro-inflammatory gene expression via inhibiting PARP-1 activity. (A) MLE-12 cells were normally cultured or stimulated with TNF-α for different times, as indicated, or (B) the cells were normally cultured or stimulated with TNF-α (in the presence or absence of PJ34 or daidzein) for 6 h. The cells were lysed, the lysates (20 μg protein) were resolved using SDS-PAGE, and protein PARylation was detected using the anti-PAR antibody. (C and D) MLE-12 cells were co-transfected with the PARP-1 expression plasmid (or the negative control pcDNA3) with the reporter plasmids NF-κB-luc or Cxcl2-luc, as indicated. After stimulation with TNF-α with or without inhibitors, a dual reporter assay was performed as described above. The transcriptional activity of NF-κB or Cxcl2 in cells transfected with the control plasmid (pcDNA3) was considered the baseline and valued as 1. (E) Cells (1×107) were stimulated with TNF-α in the presence or absence of daidzein or PJ34 for 6 h, and immunoprecipitation was performed. Antibodies against PARP-1 (left panel) or RelA/p65 (right panel) were used to obtain complexes that were resolved using SDS-PAGE. Immunoblotting was performed using antibodies specific for RelA/p65, PARP-1 and PAR, as indicated. Data (mean±SD) are representative of three independent experiments (cP<0.01 compared with cells transfected with pcDNA3; fP<0.01 compared with TNF-α-stimulated cells).
Our previous study showed that PARP-1 modulates NF-κB activity in TNF-α-stimulated murine macrophages27. To further address the inhibitory effect of daidzein on PARylation that accounted for its decreased pro-inflammatory gene expression in murine MLE-12 epithelial cells, we performed co-transfection by over-expressing PARP-1 with the NF-κB reporter plasmid. Upon stimulation by TNF-α, cells over-expressing PARP-1 revealed increased NF-κB activation compared to those transfected with pcDNA3 (approximately 3-fold). However, the presence of daidzein or PJ34 reversed this transcriptional activation, which implied that daidzein modulates PARP-1 activity and subsequently inhibits NF-κB activation (Figure 3C). Likewise, treatment with daidzein and PJ34 exhibited a similar effect on cells transfected with the Cxcl2-luc plasmid and over-expressing PARP-1 (Figure 3D).
Our previous study showed that TNF-α stimulation resulted in increased binding of PARP-1 and enhanced PARylation of RelA/p65, which is an important subunit of activated NF-κB27. To investigate the molecular mechanism by which daidzein modulates the activation of transcription factor NF-κB in the present study, we performed immunoprecipitation assays. The results showed that treatment with daidzein diminished the TNF-α-induced increase of RelA/p65 abundance in the PARP-1-associated complex and its PARylation level (Figure 3E), which suggested that daidzein modulates PARP-1 activity and impacts its binding to RelA/p65 and the transcription of NF-κB.
Daidzein modulates PARP-1 activity via direct binding
Most commercially available inhibitors or those that are already being used in clinical trials share structural similarity with the PARP-1 NAD+ substrate; therefore, they competitively block the formation of the PAR polymer11. To address whether daidzein inhibits PARP-1 activation by directly acting on PARP-1 or through another indirect mechanism, we utilized an in vitro PARP-1 activity detection kit (Trevigen). We first determined the linear range for the activity of recombinant PARP-1. The results revealed a good linear correlation for PARP-1 activity between 0.01 and 0.125 U (Figure 4A). Therefore, we further examined the effect of daidzein on PARP-1 activation using 0.1 U recombinant PARP-1. The results showed that 10 μmol/L of daidzein significantly decreased PARP-1 activity (∼75%), whereas the PARP-1 inhibitor 3-AB (10 μmol/L; provided by the kit) and PJ34 decreased the PARylation of the substrate to nearly the basal level (Figure 4B). Considering the inhibitory effect of daidzein on protein PARylation in cells exposed to TNF-α (Figure 3A), the combined results imply that Daidzein may directly bind to PARP-1. However, this activity might rely on indirect signaling mechanisms other than direct action on PARP-1 in an intracellular context.
Daidzein inhibits PARP-1 activity. PARP-1 activity assay was conducted following the instructions of the manufacturer. (A) The linear correlation intervals of PARP-1 activity with different units of recombinant PARP-1. (B) The effect of daidzein and different inhibitors on the activity of PARP-1 (0.1 U). Data (mean±SD) of each point represent the median value from triplicate experiments. Representatives of three independent experiments are shown (cP<0.01 compared with sample without inhibitor).
Discussion
Many flavones or polyphenolic flavonoids are derived from plants and play numerous pharmacological roles. These molecules possess anti-cancer properties via acting on DNA topoisomerases or vascular protection roles through regulating the cellular redox status1,2. Recently, the anti-viral, anti-microbial, anti-inflammatory and anti-allergic potential of polyphenolic flavonoids has been introduced3. Among these, daidzein, which is a natural isoflavone found in soybean and traditional medicinal herbs, shows anti-inflammatory activity and is a promising treatment option for inflammation-related diseases; however, the mechanisms for this process must be further investigated.
Daidzein pretreatment was found to significantly suppress LPS-induced mRNA expression of the pro-inflammatory factors nitric oxide and IL-6 in murine microglial cells, which suggests the neuroprotective properties of daidzein28. Release of soluble pro-inflammatory factors appears to be via inhibition of oxidative induction and activation of the p38 MAP kinase-NF-κB pathway28. Another study showed that daidzein significantly inhibited the production of NO and IL-6 and decreased the mRNA expression of NOS and IL-6 in LPS-treated RAW264.7 cells; however, the JNK and p38 pathways were not involved in daidzein-mediated suppression of NF-κB-mediated transcription6.
Recently, the anti-inflammatory activity of some food-derived flavonoids was evaluated in mouse models. The effect of these flavonoids on PARP-1 inhibition in LPS-induced, acute pulmonary inflammation was evaluated29,30. Some of these molecules, such as fisetin, significantly decreased lung myeloperoxidase levels and the gene expression of inflammatory mediators such as IL-6, TNF-α, IL-1β, MIP-1alpha, and MIP-2 (also known as Cxcl1 and Cxcl2)30. Additionally, PARP-1 activation in pulmonary epithelial cells was inhibited by fisetin treatment29.
Here, we showed that TNF-α-induced pro-inflammatory chemokine expression and inflammation in mouse lung were significantly attenuated after daidzein administration. Whether the anti-inflammatory role of daidzein, similar to fisetin, is related to its capacity to inhibit the activation of PARP-1 must be determined. PARP-1 can be activated by genotoxicity due to oxidative stress-induced DNA breaks and the extracellular signal-triggered MAPK pathway cascade12. Activated PARP-1 contributes to the pathogenesis of inflammatory diseases, which involves its regulation of gene expression via acting on various transcription factors such as NF-κB, AP-1 and Sp112. Therefore, we investigated whether daidzein inhibits the transcriptional activation of an NF-κB-containing promoter in response to TNF-α exposure.
Utilizing TNF-α-stimulated murine MLE-12 epithelial cells, we showed that PARP-1 activity was decreased by daidzein, similar to what was observed after PJ34 (a prototypic PARP-1 inhibitor) treatment. Importantly, the transcriptional activity of NF-κB and the mRNA level of Cxcl2 were also diminished in mouse lung and cultured cells. Furthermore, dual reporter assays that utilized cells overexpressing PARP-1 and an immune-precipitation assay of PARP-1-associated RelA/p65 revealed that daidzein treatment depresses PARP-1 activation. These effects of daidzein could account for the disruption of pro-inflammatory gene transcription. The results suggest the possibility that daidzein inhibits PARP-1 activity in epithelial cells, thereby controlling the exacerbation of inflammation in TNF-α-induced lungs. An in vitro PARP-1 activity assay showed that 10 μmol/L of daidzein effectively reduced PARP-1 activity. Nevertheless, this result does not exclude the possibility of daidzein depressing PARP-1 activation through another signal pathway, as was previously interpreted, because we showed that the same concentration of daidzein diminished the protein PARylation caused by TNF-α stimulation to the basal level in a cellular context. Because of the structural and natural similarity of daidzein to genistein31, we assumed that tyrosine phosphorylation might have a regulatory function in modulating PARP-1 activity. Maruyama et al reported that Txk (resting lymphocyte kinase, Rlk/Txk), which is a member of the non-receptor tyrosine kinase of Tec (tyrosine kinase expressed in hepatocellular carcinoma) family, induced PARP-1 activity, which, in turn, regulated interferon-γ gene transcription in T-helper type 1 cells32.
This work provides a rationale for further exploring the exact mechanisms by which daidzein impairs PARP-1 activity. Currently, the anti-inflammatory mechanisms of daidzein and other phytoisoflavonoids that may rationalize the scientific application of these compounds remain to be investigated. Because long-term intake of corticosteroids is accompanied by serious adverse effects, our results, combined with those of others, indicate that the phytoisoflavonoid daidzein could be a promising candidate for the treatment of chronic inflammatory diseases.
Authors contribution
Hai-yan LI, Lang PAN, Yue-shuang KE, and Enkhzaya BATNASAN conducted the experiments and collected the data; Hai-yan LI drafted the article; Zhong-ying LIU and Xue-qing BA conceived and designed the study and revised the manuscript; and Xiang-qun JIN provided materials and technical support.
Abbreviation
TNF-α, tumor necrosis factor alpha; PARP-1, poly-ADP-ribose polymerase 1; PARylation, poly-ADP-ribosylation; PAR, poly-ADP-ribose; Cxcl2, CXC-motif chemokine ligand-2; NF-κB, nuclear factor kappa B.
References
Russo P, Del Bufalo A, Cesario A . Flavonoids acting on DNA topoisomerases: recent advances and future perspectives in cancer therapy. Curr Med Chem 2012; 19: 5287–93.
Mann GE, Bonacasa B, Ishii T, Siow RC . Targeting the redox sensitive Nrf2-Keap1 defense pathway in cardiovascular disease: protection afforded by dietary isoflavones. Curr Opin Pharmacol 2009; 9: 139–45.
Chirumbolo S . The role of quercetin, flavonols and flavones in modulating inflammatory cell function. Inflamm Allergy Drug Targets 2010; 9: 263–85.
Chan YC, Wu CC, Chan KC, Lin YG, Liao JW, Wang MF, et al. Nanonized black soybean enhances immune response in senescence-accelerated mice. Int J Nanomedicine 2009; 4: 27–35.
Wei J, Bhatt S, Chang LM, Sampson HA, Masilamani M . Isoflavones, genistein and daidzein, regulate mucosal immune response by suppressing dendritic cell function. PloS One 2012; 7: e 47979.
Choi EY, Jin JY, Lee JY, Choi JI, Choi IS, Kim SJ . Anti-inflammatory effects and the underlying mechanisms of action of daidzein in murine macrophages stimulated with Prevotella intermedia lipopolysaccharide. J Periodontal Res 2012; 47: 204–11.
Schreiber V, Dantzer F, Ame JC, de Murcia G . Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7: 517–28.
Woodhouse BC, Dianov GL . Poly ADP-ribose polymerase-1: an international molecule of mystery. DNA Repair (Amst) 2008; 7: 1077–86.
Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A 2006; 103: 18308–13.
D'Amours D, Desnoyers S, D'Silva I, Poirier GG . Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 1999; 342: 249–68.
Jagtap P, Szabo C . Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov 2005; 4: 421–40.
Ba X, Garg NJ . Signaling mechanism of poly(ADP-ribose) polymerase-1 (PARP-1) in inflammatory diseases. Am J Pathol 2011; 178: 946–55.
Ba X, Gupta S, Davidson M, Garg NJ . Trypanosoma cruzi induces the reactive oxygen species-PARP-1-RelA pathway for up-regulation of cytokine expression in cardiomyocytes. J Biol Chem 2010; 285: 11596–606.
Mazzon E, Genovese T, Di Paola R, Muia C, Crisafulli C, Malleo G, et al. Effects of 3-aminobenzamide, an inhibitor of poly (ADP-ribose) polymerase, in a mouse model of acute pancreatitis induced by cerulein. Eur J Pharmacol 2006; 549: 149–56.
von Lukowicz T, Hassa PO, Lohmann C, Boren J, Braunersreuther V, Mach F, et al. PARP1 is required for adhesion molecule expression in atherogenesis. Cardiovasc Res 2008; 78: 158–66.
Chiang J, Shen YC, Wang YH, Hou YC, Chen CC, Liao JF, et al. Honokiol protects rats against eccentric exercise-induced skeletal muscle damage by inhibiting NF-kappaB induced oxidative stress and inflammation. Eur J Pharmacol 2009; 610: 119–27.
Jog NR, Dinnall JA, Gallucci S, Madaio MP, Caricchio R . Poly(ADP-ribose) polymerase-1 regulates the progression of autoimmune nephritis in males by inducing necrotic cell death and modulating inflammation. J Immunol 2009; 182: 7297–306.
Park EM, Cho S, Frys K, Racchumi G, Zhou P, Anrather J, et al. Interaction between inducible nitric oxide synthase and poly(ADP-ribose) polymerase in focal ischemic brain injury. Stroke 2004; 35: 2896–901.
Khan AQ, Khan R, Rehman MU, Lateef A, Tahir M, Ali F, et al. Soy isoflavones (daidzein & genistein) inhibit 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced cutaneous inflammation via modulation of COX-2 and NF-kappaB in Swiss albino mice. Toxicology 2012; 302: 266–74.
Park MH, Ju JW, Park MJ, Han JS . Daidzein inhibits carbohydrate digestive enzymes in vitro and alleviates postprandial hyperglycemia in diabetic mice. Eur J Pharmacol 2013; 712: 48–52.
Ba X, Bacsi A, Luo J, Aguilera-Aguirre L, Zeng X, Radak Z, et al. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J Immunol 2014; 192: 2384–94.
Hasumuma R, Kawaguchi K, Kikuchi S, Sugiyama T, Kumazawa Y . Effects of isoflavones and soybeans fermented with Bacillus subtilis on lipopolysaccharide-induced production of tumor necrosis factor-alpha and fibrinolysis in vivo. Immunopharm Immunot 2007; 29: 323–33.
Mota R, Sánchez-Bueno A, Saenz F, Hernández-Espinosa L, Jimeno D, Tornel J, et al. Inhibition of poly(ADP-ribose) polymerase attenuates the severity of acute pancreatitis and associated lung injury. Lab Invest 2005; 85: 1250–62.
Gras D, Chanez P, Vachier I, Petit A, Bourdin A . Bronchial epithelium as a target for innovative treatments in asthma. Pharmacol Ther 2013; 140: 290–305.
Lambrecht BN, Hammad H . The airway epithelium in asthma. Nat Med 2012; 18: 684–92.
Proud D, Leigh R . Epithelial cells and airway diseases. Immunol Rev 2011; 242: 186–204.
Liu L, Ke Y, Jiang X, He F, Pan L, Xu L, et al. Lipopolysaccharide activates ERK-PARP-1-RelA pathway and promotes nuclear factor-kappaB transcription in murine macrophages. Hum Immunol 2012; 73: 439–47.
Chinta SJ, Ganesan A, Reis-Rodrigues P, Lithgow GJ, Andersen JK . Anti-inflammatory role of the isoflavone daidzein in lipopolysaccharide-stimulated microglia: implications for Parkinson's disease. Neurotox Res 2013; 23: 145–53.
Geraets L, Moonen HJ, Brauers K, Wouters EF, Bast A, Hageman GJ . Dietary flavones and flavonoles are inhibitors of poly(ADP-ribose)polymerase-1 in pulmonary epithelial cells. J Nutr 2007; 137: 2190–5.
Geraets L, Haegens A, Brauers K, Haydock JA, Vernooy JH, Wouters EF, et al. Inhibition of LPS-induced pulmonary inflammation by specific flavonoids. Biochem Biophys Res Commun 2009; 382: 598–603.
Moutsatsou P . The spectrum of phytoestrogens in nature: our knowledge is expanding. Hormones (Athens) 2007; 6: 173–93.
Maruyama T, Nara K, Yoshikawa H, Suzuki N . Txk, a member of the non-receptor tyrosine kinase of the Tec family, forms a complex with poly(ADP-ribose) polymerase 1 and elongation factor 1alpha and regulates interferon-gamma gene transcription in Th1 cells. Clin Exp Immunol 2007; 147: 164–75.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31371293), the Natural Science Foundation of Jilin Province in China (201015108) and the International Science-Technology Collaboration Foundation of Jilin Province in China (20120728). We thank Dr Patrick A ZWEIDLER-MCKAY (The University of Texas MD Anderson Cancer Center) for his generous gift of PARP-1 plasmid.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Li, Hy., Pan, L., Ke, Ys. et al. Daidzein suppresses pro-inflammatory chemokine Cxcl2 transcription in TNF-α-stimulated murine lung epithelial cells via depressing PARP-1 activity. Acta Pharmacol Sin 35, 496–503 (2014). https://doi.org/10.1038/aps.2013.191
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.191
Keywords
This article is cited by
-
Isoflavones daidzin and daidzein inhibit lipopolysaccharide-induced inflammation in RAW264.7 macrophages
Chinese Medicine (2022)
-
PARP1 promotes gene expression at the post-transcriptional level by modulating the RNA-binding protein HuR
Nature Communications (2017)
-
Strategic questions about China’s shale gas development
Environmental Earth Sciences (2015)