Abstract
Aim:
Cancer stem cells have the capacity to initiate and sustain tumor growth. In this study, we established a CD44+ colorectal cancer stem cell line with particular emphasis on its self-renewal capacity, enhanced tumor initiation and drug resistance.
Methods:
Fresh colon cancer and paired normal colon tissues were collected from 13 patients who had not received chemotherapy or radiotherapy prior to surgery. Among the 6 single-cell derived clones, only the P6C cell line was cultured for more than 20 passages in serial culture and formed holoclones with high efficiency, and then the stemness gene expression, colony formation, tumorigenicity and drug sensitivities of the P6C cell line were examined.
Results:
Stemness proteins, including c-Myc, Oct3/4, Nanog, Lgr5, and SOX2, were highly expressed in the P6C cell line. Oct3/4-positive P6C cells mostly generated holoclones through symmetric division, while a small number of P6C cells generated meroclones through asymmetric division. P6C cells stably expressed CD44 and possessed a high capacity to form tumor spheres. A single cell-derived sphere was capable of generating xenograft tumors in nude mice. Compared to SW480 and HCT116 colorectal cancer cells, P6C cells were highly resistant to Camptothecin and 5-fluorouracil, the commonly used chemotherapeutic agents to treat colorectal cancers.
Conclusion:
We established a colorectal cancer stem cell line P6C with a high tumorigenic capacity and the characteristics of normal stem cells. It will benefit the mechanistic studies on cancer stem cells and the development of drugs that specifically target the cancer stem cells.
Similar content being viewed by others
Introduction
The cancer stem cell (CSC) hypothesis provides a new insight into understanding tumor initiation, recurrence and metastasis. According to this theory, tumors are organized in a hierarchy of heterogeneous cell populations, and only a small subset of cells, namely, the CSCs or tumor-initiating cells, possesses the ability to drive and sustain tumor growth1,2. The presence of CSCs in neoplastic tissue has long been hypothesized, and recently, these cells have been identified3,4. CSCs were first identified in leukemia, but they have since been identified in solid tumors. A variety of putative cell surface markers, including CD34, CD44, CD133, CD24, ALDH, and Lgr5, have been reported5,6,7,8,9. The freshly isolated CSCs or tumor initiating cells displayed enhanced tumorigenicity and could reconstruct the original tumor when transplanted into immunodeficient mice. CSCs are proposed to have stem cell properties; they are capable of undergoing extensive proliferation and self-renewal through symmetric division and differentiation into non-tumorigenic cancer cells through asymmetric division. The true signatures or markers for CSCs have yet to be identified, although there are reports suggesting that both CSCs and normal stem cells express key transcription factors, including Oct3/4, Nanog and SOX210.
Sufficient amounts of CSCs that share the same background and stable phenotypes are needed to perform reliable functional assays. However, CSCs isolated from patients are generally rare and likely undergo differentiation during maintenance in culture, resulting in a shortage of material for mechanistic studies or for screening new drugs specific to CSCs. Most CSC assays depend on the enrichment of CSCs from freshly isolated tumors, and the efficiency of cell sorting and the varied genetic background can also hamper the research into CSCs. The contamination of freshly sorted CSCs with lymphocytes or stoma cells can influence the subsequent analysis. Thus, the establishment of human colorectal CSC lines is an enticing strategy to investigate the mechanisms of tumor initiation, drug resistance, metastasis and recurrence. Until now, although more than 50 colorectal cancer cell lines have been reported, no stable CSC lines have been established. Earlier work in our laboratory showed that the CD44+ subpopulations of human colorectal cancer cells possessed higher tumorigenicity, as well as spheroid and holoclone formation capacity, which are hallmarks of CSCs. In this study, we present a newly established CD44+ colorectal CSC line. This cell line was thoroughly examined, with a particular emphasis on its self-renewal capacity, enhanced tumor initiation, and drug resistance. This CSC line will be an invaluable resource for various future studies, including the high throughput screening of drugs and antibodies, gene manipulation, and long-term in vivo assays.
Materials and methods
Patients, animals, and cell lines
Fresh colon cancer tissues and the paired normal colon tissues were collected from the tumor bank of the Beijing Cancer Hospital (Beijing, China), as approved by the Research Ethics Board at the Beijing Institute for Cancer Research.
Four-week-old female nude mice (BALB/c-nu /nu) were purchased from the Chinese Academy of Medical Sciences, and all experiments were performed under standard conditions in accordance with the institutional regulations.
The SW480, HCT116, and HT29 colorectal cancer cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS, Hyclone) in a humidified incubator at 37 °C.
Reagents
The antibodies used in this study include the following: anti-CD31 (C20, Santa Cruz), anti-CD44 (2C5, R&D), anti-CD44-FITC (G44-26, BD PharMingen), anti-CD45-FITC (2D-1), anti-CD133 (AC133, Miltenyi), anti-cytokeratin 1 (CK1; N-20, Santa Cruz), anti-cytokeratin 20 (CK20; Ks20.8, DAKO), anti-CDX2 (AMT28, ZSBio), anti-c-Myc (9E10, Santa Cruz), anti-GFP (B-2, Santa Cruz), anti-Oct3/4 (C-10, Santa Cruz), anti-Nanog (polyclonal, R&D), anti-EpCAM (158206, R&D), anti-Lgr5-Dylight 488 (Novas), anti-SOX2 (245610, R&D), anti-ABCG2 (BXP-21, Calbiochem), and anti-CXCR4 (C-20, Santa Cruz). TRIzol and the reverse transcriptase kit were purchased from Invitrogen, and all other reagents were purchased from Sigma unless otherwise specified.
Flow cytometry
Flow cytometry was performed as previously described11. Briefly, the cells were dissociated into a single cell suspension by trypsin digestion. After being washed with PBS, the cells were incubated with antibodies at 4 °C for 20 min before flow cytometric analysis.
Karyotypic assay
Metaphase spreads for the FISH experiments were prepared according to a standard protocol12. Briefly, the cells were treated with colchicine before being fixed and stored at -20 °C. The cell suspension was dropped onto cold, humidified microscope slides before M-FISH analysis. Whole chromosome painting probes used for the M-FISH analysis have been described in a previous study13, and the FISH assay was performed as described previously12.
Cell proliferation assays
The cells were cultured in 6-well plates and were then trypsinized and resuspended in 200 μL PBS. The viable cells were counted every day under the microscope following trypan blue staining. For cell cycle analysis, 1×106 cells were harvested and fixed with 75% ethanol at -20 °C; the cells were then incubated with 50 μg/mL PI and 1 mg/mL RNase A for 30 min. The DNA content was measured using a flow cytometer.
Sphere formation and colony formation assays
For the sphere formation assay, the cells were transplanted into a 6-well plate pre-coated with a thin layer of 1.2% agar at a density of 100 cells per well. Spheres or spheroids that arose within 3 weeks were observed and counted. The samples were analyzed in triplicate for each cell type, and at least three independent experiments were carried out.
For the colony formation assay, a single cell suspension was obtained through trypsinization and filtration through a 40-μm filter; the cells were then transplanted into a 6-well plate containing 0.35% soft agar at a concentration of 100 cells per well. Colonies were observed under a phase contrast microscope and stained with crystal violet on d 20. Each sample was analysed in triplicate, and this experiment was performed three times.
Immunofluorescence staining
The cells grown on coverslips were fixed with 3.7% paraformaldehyde and permeabilized with 0.2% Triton X-100. After incubation with the primary antibody for 1 h, a fluorochrome-conjugated secondary antibody was added for 45 min. The cells were washed completely with PBS, then mounted and observed under a fluorescent microscope.
Tumorigenicity assay
To determine the cell line's tumor formation ability, 100 to 2×106 cells, or a single cell–derived sphere, were injected sc into 4.5-week-old nude mice (6–8 mice per group). Tumors were measured using a slide gauge every 3 d, and the tumor volume was calculated as 1/2×length×width2. The tumor latency and incidence were recorded for 90 d following transplantation.
RNA interference (RNAi)
The recombinant lentivirus-pseudotyped particles were generated using a triple plasmid system as previously described11. Briefly, the 293T cells were co-transfected with 3 plasmids to package pseudo-lentiviruses. Stably infected cells were isolated following puromycin screening, and the RNA interference efficiency was determined using Western blot analysis.
Western blotting
The cells were fractionated in lysis buffer (10 mmol/L HEPES, pH 7.4, 2 mmol/L EGTA, 0.5% Nonidet P-40, 1 mmol/L NaF, 1 mmol/L NaVO4, 1 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L dithiothreitol, 50 μg/mL trypsin inhibitor) on ice for 30 min. Equivalent samples were subjected to 12% SDS-PAGE and were then transferred onto nitrocellulose membranes. The membranes were incubated with the indicated antibodies, and immunoreactive bands were visualized using enhanced chemiluminescence (Pierce).
Chemosensitivity assay
To determine the cell line's sensitivity to chemotherapy, 2×106 cells were cultured in 6-well plates for 24 h and were then treated with camptothecin or 5-FU for 24, 48, or 72 h. Cell death was detected through bright field microscopic observation and flow cytometric analysis following Annexin V/PI staining.
Results
Establishment of CD44-positive colorectal cancer cell lines
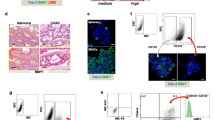
We collected fresh colon cancer tissues and paired normal colon tissues from 13 patients who had not received chemotherapy or radiotherapy prior to surgery. The tissues were minced in DMEM and were incubated with 1 mg/mL collagenase and 1 mg/mL hyaluronidase for 1 h. The cell suspension was plated in 25-cm2 flasks, containing DMEM supplemented with 10% FBS, and ultra-low attachment dishes containing DMEM supplemented with 5% FBS. Colorectal cancer cells from eight patients generated spheres in the ultra-low attachment dishes (Corning, #3262) after 25 d (Figure 1A). In contrast, under identical culture conditions, the cells isolated from the paired normal colon tissues did not form any spheres. Instead, the normal colon cells underwent limited cell division before senescing (Figure 1A). We then isolated single cancer cells from the spheres and implanted these cells at a concentration of 0.5 cells per well in a 96-well plate. Six single-cell derived clones (named P6C, P7C, P8C, P13C-1, P13C-2, and P13C-3) were generated from four patients. Among them, only the P6C line was cultured for more than 20 passages in serial culture and formed holoclones with high efficiency14,15 (Figure 1A).
Characterization of the P6C cells. (A) Holoclone formation of the P6C cells. Cells from primary colorectal cancer tissue and paired normal colon tissue were trypsinized and seeded into a 6-well plate. A primary clone from a single cancer cell and a holoclone of the P6C cells (passage 20) are shown in the top column, respectively. A representative primary sphere from the colon cancer tissue and dispersed normal colon cells are shown in the bottom column. Scale bars, 200 μm. (B) Expression of distinct markers in the P6C cells. Suspended P6C cells were incubated with anti-CD45-FITC, CD31-FITC, and CD24-FITC, respectively, and were analysed by flow cytometry. Donkey anti-mouse-FITC was used as a control. M1, negative; M2, positive. (C) Surface expression of CD44 in P6C cells as detected by immunofluorescence. P6C cells were grown attached to plates for 5 d, fixed with paraformaldehyde and incubated with anti-CD44 antibody. DAPI was used to stain the nucleus. Scale bar, 100 μm. (D) Relative expression levels of distinct markers under different culture conditions, including a P6C sphere, a P6C clone and SW480 cells. Suspended cells were collected and incubated with FITC conjugated antibodies and were analysed by flow cytometry. Each sample was analysed in triplicate, and the experiment was repeated 3 times. bP<0.05, cP<0.01. (E) Comparison of the clonal formation of the P6C, HCT116, and SW480 cell lines. A small number of cells (200) were cultured in 6-well plates for 20 d, and the resulting clones were stained with crystal violet (left). The results of the statistical analysis are shown in the right column. Each cell was seeded in triplicate, and the experiment was repeated 3 times. cP<0.01.
The P6C cell line was derived from a sigmoid colon cancer from a 48-year-old male with concurrent lymph nodes metastases and has been sub-cultured in serum-free DMEM in ultra-low attachment dishes for more than 120 passages, thereby becoming an immortalized cell line. To confirm its epithelial derivation, we examined the expression of several types of surface markers, including CD24, CD31, CD45, and EpCAM, in the P6C cells. Flow cytometric analysis showed that P6C cells expressed the epithelial marker CD24, but not the leukocyte markers CD31 or CD45 (Figure 1B; Supplementary Figure 1). Immunofluorescence staining confirmed that P6C cells were EpCAM+ (data not shown). Interestingly, we determined that P6C cells express CD44, a well-known colorectal CSC marker (Figure 1C; Supplementary Figure 2). It was reported that when the cells are allowed to grow in sphere cultures, they display more CSC properties; thus, we examined the CD44 expression level in the P6C cell line under different culture conditions. Flow cytometric analysis showed that ∼99% of the P6C cells in spheres expressed CD44, and this percentage decreased to ∼90% when cells were grown attached to plates (Figure 1D). In contrast, the expression levels of differentiated intestinal epithelial markers, including CDX2, cytokeratin 1 (CK1) and CK20, were low when cells were grown under spheroid culture conditions and increased significantly upon attachment. This suggests that P6C cells in spheroids maintain an undifferentiated status and undergo a certain degree of differentiation when cultural conditions change (Figure 1D). When compared with the P6C cell line, the differentiated colorectal cancer cell line SW480 exhibited low expression of CD44, high expression of CK20 and was positive for CDX2 expression (Figure 1D).
Enhanced clonal formation is one of the characteristics of CSCs. When 100 cells were implanted into a 6-well plate, we observed that a larger number of clones were generated from the P6C cell line than from the HCT116 and SW480 cell lines; this difference in clone number was significant (Figure 1E). These data indicate that the P6C cell line has the capacity to undergo self-renewal and differentiation, both of which are characteristics of stem cells.
P6C cells express stemness genes
We next asked if the CD44-positive P6C cells, which can grow in spheroids in ultra-low attachment dishes, express stemness genes. By immunofluorescence staining, we observed that c-Myc, Oct3/4, Nanog and SOX2, markers of normal stem cells, were all expressed in P6C cells (Figure 2A). Western blotting analysis further confirmed that Oct3/4 was highly expressed in P6C cells (Figure 2B). In addition, the normal colon stem cell marker Lgr5 was also detected in P6C cells by Western blotting (Figure 2B). In contrast, these stemness transcription factors were undetectable in the HT29, HCT116, and SW480 cell lines, which are differentiated colorectal cancer cell lines (Figure 2B). Because our previous data showed that CD44 is of functional importance for CSCs, we next addressed whether the expression of CD44 could affect the stemness of P6C cells. To this end, we stably depleted cells of CD44 through specific shRNA inhibition and found that Oct3/4 expression was significantly decreased in CD44-depleted cells (Figure 2C, Supplementary Figure 3 and 4). In a well-established holoclone, immunofluorescence staining showed that Oct3/4 and CD44 were co-expressed in the central region of the clone, while the Oct3/4 and CD44 double negative differentiated cancer cells localized to the outer surface of the clone (Figure 2D). Taken together, these data indicated that P6C cells share transcription factors with normal stem cells.
Stemness genes expression in P6C cells. (A) Immunofluorescence of stemness proteins in cultured P6C cells. P6C spheres were dissociated, seeded onto coverslips and allowed to attach for 3 h. After fixing cells with paraformaldehyde, the cells were incubated with the indicated antibodies. DAPI was used for nuclei counter staining. Scale bars, 100 μm. (B) Stemness gene expression detected by Western blotting. P6C cells were stably transfected with the pOct3/4 promoter-EGFP (OPG) construct. The whole cell lysates of HT29, HCT116, SW480, P6C, and P6C-OPG cells were loading equally, subjected to SDS-PAGE and transferred to nitrocellulose membranes. The membranes were then incubated with the indicated antibodies and visualized using an ECL system. (C) Effect of CD44 shRNA on Oct3/4 expression in P6C cells. Relative Oct3/4 promoter activity was measured by GFP fluorescence intensity as detected using flow cytometry. P6C and parental cells were used as controls. Each sample was performed in triplicate, and the experiment was repeated 3 times. bP<0.05. (D) Co-expression of endogenous Oct3/4 and CD44 in a P6C single-cell derived clone. Magnified images are shown in the lower right panel. Scale bars, 50 μm.
Self-renewal and differentiation of P6C cells
By definition, CSCs should display the stem cell properties of both symmetric and asymmetric divisions, which are critical for self-renewal and differentiation, respectively. To address this issue, we stably inserted EGFP downstream of the endogenous Oct3/4 promoter in both the P6C and SW480 cell lines (pOct3/4 promoter-EGFP, OPG, a gift from Dr Ying JIN). Flow cytometric analysis revealed that more than 95% of the P6C cells in holoclones expressed Oct3/4, while less than 5% of the SW480 cells expressed Oct3/4 (Figure 2B and 3A).
Symmetric and asymmetric division of P6C cells. (A) Oct3/4 promoter activation in P6C and SW480 cells. P6C and SW480 cells were stably transfected with the pOct3/4 promoter-EGFP construct. Oct3/4 promoter activation and CD45 expression were measured by fluorescence intensity using a flow cytometer. (B) Self-renewal of an Oct3/4 positive P6C cell. A single Oct3/4 positive P6C cell was seeded in a 6-well plate and observed under a microscope. Pictures were taken every 24 h to detect holoclone formation. Scale bars, 50 μm. (C) Asymmetric division of an Oct3/4 positive P6C cell. A meroclone from a single Oct3/4 positive P6C cell underwent asymmetric division at the 2-cell stage. Pictures were taken every 24 h. Scale bars, 50 μm. (D) Distinct meroclone, paraclone and holoclones derived from Oct3/4 positive cells. Cell morphology in an Oct3/4 partial positive clone (top) showed the tight adhesion of the Oct3/4-positve cells and the loosen interaction of the Oct3/4-negative cells. Two side-by-side clones indicating that the paraclone is Oct3/4-negative and the holoclone is Oct3/4-positive (bottom). Scale bars, 200 μm.
Self-renewal and differentiation are hallmarks of stem cells. To understand their roles in the P6C cell line, we implanted single P6C-OPG cells into a 96-well plate and observed their division under the microscope. The majority of the Oct3/4-positive cancer cells underwent symmetric division and formed GFP-positive holoclones cells (87%), as shown in Figure 3B, whereas only 13% of the P6C cells generated meroclones through asymmetric division (Figure 3C). A small percentage (∼0.5%) of Oct3/4-negative cells were able to de-differentiate into Oct3/4-positive cells when seeded in ultra-low attachment dishes (data not shown). We also noticed that Oct3/4 expression was highly correlated with cell morphology. All of the paraclones were Oct3/4-negative, whereas all of the observed holoclones consisted of Oct3/4-positive cells. In a meroclone, the cells expressing Oct3/4 adhered tightly, while the Oct3/4-negative cells were loosely contacted, as observed under a phase-contrast microscope (Figure 3D). These observations further support the hypothesis that P6C cells have the critical properties of the self-renewal and differentiation. In addition, Oct3/4 can be a marker of colorectal CSCs, which have greater potential to undergo symmetric division as previously suggested16.
Chromosomal instability and mutations in p53 in P6C cells
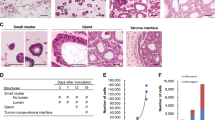
One key feature of a cancer cell versus a normal cell is chromosomal instability, which is proposed to be critical for the initiation of tumorigenesis17; however, the exact role that genomic instability plays in the initiation of CSCs remains elusive. Thus, we were interested to investigate the genomic integrity of the P6C cell line. As shown in Figure 4A, 73% of the P6C cells possessed 59 chromosomes. There was one copy of chromosomes 3, 4, 9, 13, 14, and 15 and three copies of chromosomes 7, 12, 20, and 22. Chromosomal translocations were also frequent in this cell line, along with chromosomal insertions and deletions (Figure 4A). These data reveal that the P6C cell line suffered from chromosomal instability and abnormal mitosis, both common features of cancer cells.
Karyotype and p53 mutations of P6C cells. (A) Representative karyotype of the P6C cells. Approximately 73% of the P6C cells possessed 59 chromosomes. Every chromosome was stained with a different color through FISH analysis. (B) The 72P to 72R mutation in a p53 allele of the P6C cells (passage 120). p53 cDNA was reverse transcribed from mRNA and cloned into a T-easy vector prior to sequencing (top). The C to G mutation was confirmed using Chromos Map (bottom). (C) A 117-bp insertion in the p53 cDNA from P6C cells, which resulted in a premature “TGA” stop codon after 25 amino acids. (D) p53 alleles from passage 4 and passage 120 P6C cells. The cDNA was reverse transcribed from mRNA, and the p53 gene was amplified by PCR before being inserted into the T-vector. The statistical analysis of the p53 alleles was performed by sequencing 10 to 22 constructs from each sample.
Genetic mutation of tumor suppressor genes, such as p53, has been closely associated with the initiation of cancers. We were interested in whether CSCs have a mutated p53 gene, which may be related to their abnormal proliferation. The p53 gene was cloned from the P6C cell line, and sequencing analysis revealed that 72P to R mutants occurred in 60% and 67% cells of passage 4 and 120, respectively (Figure 4B, 4D). Additionally, we found a 117 bp insertion in the p53 cDNA; this insertion resulted in a truncated 25 amino acids at the N-terminal of p53 (Figure 4C). Importantly, we found that the mutations in the p53 gene were similar in both low passage cells (passage 4) and high passage cells (passage 120), strongly suggesting that these mutations did not accumulate due to the in vitro cell culture conditions (Figure 4D). These data also support the possibility that mutations in certain stem cells could lead to the occurrence of CSCs, as previously proposed.
We also determined the proliferation rate of the P6C cells in monolayer culture by calculating the cell growth rate. As shown in Supplementary Figure 5, the doubling time of P6C cells was ∼20 h, similar to the SW480 and HCT116 cell lines (P>0.05). These data indicated that P6C cells share similar proliferation properties with differentiated colorectal cancer cell lines when grown attached to cell culture dishes.
Xenograft tumor from a single cell-derived holoclone
We next addressed whether a single P6C cell could result in a xenograft tumor, a distinguishing feature of CSCs. Single cell-derived holoclones containing approximately 500 cells were injected into nude mice. Strikingly, xenograft tumors initiated with 100% incidence (6/6). We then dissociated and trypsinized the tumors from the nude mice to regenerate secondary single cell-derived clones. Re-injection of the secondary clones, approximately 500 cells, into nude mice also resulted in 100% tumor initiation incidence. These data also support the notion that P6C cells possess the capacity for self-renewal and differentiation in vivo.
One advantage of a CSC line is to provide an ideal system to trace tumor development in vivo. For this strategy, we pre-labeled P6C-OPG cells with DsRed and purified the double positive cells using FACS (Figure 5A). DsRed/GFP double positive holoclones were selected and transplanted into nude mice (Figure 5B). Tumor development was visualized by whole body fluorescence imaging, as shown in Figure 5C. Immunohistochemistry of GFP revealed that certain Oct3/4-positive P6C cells differentiated into Oct3/4-negative cells in the xenograft tumors. The co-localization of CD44 and Oct3/4 was detected by serial section staining. Interestingly, CD44 and Oct3/4 double positive cells resided in clusters in the xenograft tumors, similar to primary cancers (Figure 5D).
Imaging of tumors derived from a single cell derived clone of P6C. (A) DsRed labeled P6C-OPG cells were sorted using FACS. P6C-OPG cells were transfected with the DsRed construct and analysed by FACS. The GFP/DsRed double positive cells were separated out for propagation in culture dishes. (B) GFP/DsRed double positive clones. The DsRed was stably expressed in a single cell derived from a P6C-OPG clone. Scale bars, 200 μm. (C) Xenograft tumors initiated from the DsRed labeled P6C-OPG holoclones in nude mice. Tumors were detected by whole body fluorescent imaging (in-vivo FX PRO, Carestream) and were shown by relative fluorescence intensity at d 0 and 30. (D) Immunohistochemistry of xenograft tumors. Tumors were fixed in paraffin, and CD44 and GFP antibodies were used to detect CD44 and Oct3/4 expression. Scale bars, 200 μm.
Drug resistance of the P6C cells
It has been proposed that CSCs are able to confer drug resistance and contribute to cancer recurrence. We thus sought to address the question whether the P6C cells were resistant to chemotherapeutic agents. Camptothecin (CPT) and 5-fluorouracil (5-FU) are commonly used chemotherapeutic drugs in the treatment of colorectal cancer. Compared to HCT116 and SW480 cells, we found that the P6C cells were less sensitive to CPT and 5-FU (Figure 6A, 6B; Supplementary Figure 6A). In addition to the lack of cell proliferation inhibition, P6C cells were highly resistant to 5-FU induced apoptosis, as shown by Annexin V/PI staining (Figure 6B, 6C; Supplementary Figure 6B). Because the most commonly used chemotherapeutic drugs function through cell cycle arrest, we thus investigated whether 5-FU affected the cell cycle. To our surprise, when compared to SW480 cells, P6C cells were less sensitive to the 5-FU-induced S-G2 checkpoint at 24 h. Although cell cycle arrest continued through 48 h, this effect was attenuated by 72 h (Figure 6D). These data indicated that there is a unique cell cycle checkpoint in CSCs, which results in drug resistance.
Chemoresistance of the P6C cells. (A) Morphological observation of P6C, HCT116 and SW480 cells treated with 2 μmol/L camptothecin. Cell morphology was observed under a microscope at 24 and 48 h. Scale bars, 100 μm. (B) 5-FU induced cell death of P6C and SW480 cells. Cells were treated with 1 μg/mL 5-FU for the indicated times and were then trypsinized. After staining with Annexin V and PI, flow cytometric analysis was performed. (C) Statistical analysis of 5-FU induced cell death. P6C and SW480 cells were treated with 0.1, 1, and 10 μg/mL 5-FU for the indicated times. Cell death was calculated as the percentage of PI+ cells, as determined by flow cytometry. This experiment was repeated 3 times. cP<0.01. (D) Cell cycle analysis of P6C cells treated with 5-FU. The DNA content of the indicated cells was detected by PI staining followed by flow cytometric analysis. Each sample was performed in triplicate, and the experiment was repeated 3 times.
Discussion
In the present study, we established a colorectal cancer cell line, P6C, which possesses all of the characteristics of CSCs. Firstly, P6C cells are CD44 positive. CD44 is one of the most widely used putative surface markers for CSCs in breast, prostate, pancreatic and colorectal cancers, and P6C cells in spheres or monoclones express high levels of CD44. Secondly, we found that stemness genes, including Oct3/4, Nanog and SOX2, are expressed in P6C cells and are absent in differentiated cancer cell lines. As master regulators of pluripotency that control lineage commitment during early development, they are key components in transforming cells into induced pluripotent stem cells18,19,20. A previous report also showed that misexpression of the OCT4 gene resulted in pronounced intestinal dysplasia21. Thirdly, we demonstrated that single cell-derived spheres or holoclones resulted in 100% xenograft tumor initiation incidence in nude mice. Finally and importantly, we showed that P6C cells could undergo self-renewal and differentiation, both functions are critical for CSCs to progress into a tumor.
It is arguable whether these cells are truly CSCs because they have not been clearly shown to undergo symmetric or asymmetric division. Using an Oct3/4 promoter driven GFP system, we demonstrated that Oct3/4-positive cells underwent both symmetric and asymmetric division to generate Oct3/4-positive and Oct3/4-negative cells to maintain colorectal CSCs in a stable proportion. Importantly, we found that Oct3/4-positive cells formed holoclones, which could regenerate xenograft tumors in nude mice, while ∼10% of the Oct3/4-positive cells underwent asymmetric division to form meroclones or paraclones. These data revealed that these CSCs could undergo both asymmetric and symmetric division as normal stem cells. Interestingly, less than 1% of the Oct3/4-negative P6C cells could become Oct3/4-positive. Although it is not clear how the Oct3/4-negative cell achieves this, de-differentiation or asymmetric division might be involved. Oct3/4 is a master regulator of stem cell properties and is involved in the regulation of symmetric division22. Indeed, we found that Oct3/4-negative daughter cells do not form holoclones and have a tendency to differentiate. Additionally, P6C cells tended to differentiate when grown in attachment, similar to normal stem cells, or to differentiate into other types of tumor cell in a xenograft model system. It is likely that cell-to-cell contact and other niche factors play a role in the regulation of asymmetric division. We found that expression of CD44 and Oct3/4 were highly correlated. Cells expressing both proteins localized in the holoclones, but not in the paraclones, and in the center of the spheres (Figure 2D). CD44 knockdown reduced the expression of Oct3/4. Further studies are needed to address whether CD44 is involved in the regulation of symmetric versus asymmetric division. Nevertheless, our data suggest that normal and cancer stem cells share the same potential for self-renewal and differentiation, although CSCs have a greater potential for self-renewal.
Genomic instability is a defining characteristic of cancer cells. However, how genomic instability is causally related to the origin of CSCs is not fully understood. It has been suggested that CSCs may arise from normal stem cells following the accumulation of genomic mutations. However, this has not been clearly demonstrated. We found that P6C cells had gross chromosomal changes, including triplicates, translocations and deletions. Mutations in the p53 gene have been reported to compromise cell death, thus promoting tumorigenesis23. Despite the fact that the P6C cell line originated from a single cell, we still detected an arginine allele occurring at a polymorphic codon 72 (72R) in the 67% of the P6C cells; this is consistent with the report that 72R is more common than 72P in cancer24. Importantly, we found that early passage cells also had similar p53 mutations. These data indicate that genomic instability occurs early during the initiation of cancer, and CSCs indeed have accumulated genomic changes. Compared to the HCT116 and SW480 cell lines, P6C cells showed a significantly enhanced resistance to camptothecin and other chemotherapeutic agents (staurosporine, As2O3 and phenylarsine oxide; data not shown). We did not detect high expression of ABCG2 or other ABC family proteins (data not shown). It is possible that p53 mutations may contribute to the drug resistance of CSCs. Consistently, patients whose cancers contain the 72R variant of the p53 gene have a worse response to therapy than those expressing the 72P variant of the p53 gene25. The mechanism by which CSCs originate from other types of cells and how these cells acquire drug resistance need to be critically evaluated in future studies.
Author contribution
Guan-hua RAO carried out the immunoassays and participated in the cell line establishment with the assistance of Xiao-hui WANG, Jun WANG, and Hai-jing JIN; Hong-min LIU carried out the cellular studies, participated in the cell culture and drafted the manuscript; Bao-wei LI and Yan-lei YANG collected the tumor samples, carried out the tumorignecity studies; Jia-jie HAO and Ming-rong WANG carried out the genetic studies; Quan CHEN designed the experiments and drafted the manuscript; and Lei DU performed the molecular studies and statistical analysis, participated in experimental design and drafted the manuscript.
References
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells — perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 2006; 66: 9339–44.
Ailles LE, Weissman IL . Cancer stem cells in solid tumors. Curr Opin Biotechnol 2007; 18: 460–6.
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994; 367: 645–8.
Bonnet D, Dick JE . Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 1997; 3: 730–7.
Maitland NJ, Collins AT . Prostate cancer stem cells: a new target for therapy. J Clin Oncol 2008; 26: 2862–70.
Dontu G, Liu S, Wicha MS . Stem cells in mammary development and carcinogenesis: implications for prevention and treatment. Stem Cell Rev 2005; 1: 207–13.
Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003; 63: 5821–8.
Ponti D, Zaffaroni N, Capelli C, Daidone MG . Breast cancer stem cells: an overview. Eur J Cancer 2006; 42: 1219–24.
Schepers AG, Snippert HJ, Stange DE, Born M, Es JH, Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 2012; 337: 730–5.
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, et al. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res 2010; 70: 10433–44.
Du L, Wang H, He L, Zhang J, Ni B, Wang X, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 2008; 14: 6751–60.
Yang Y, Chu J, Wu Y, Luo M, Xu X, Han Y, et al. Chromosome analysis of esophageal squamous cell carcinoma cell line KYSE 410-4 by repetitive multicolor fluorescence in situ hybridization. J Genet Genomics 2008; 35: 11–6.
Wu YP, Yang YL, Yang GZ, Wang XY, Luo ML, Zhang Y, et al. Identification of chromosome aberrations in esophageal cancer cell line KYSE180 by multicolor fluorescence in situ hybridization. Cancer Genet Cytogenet 2006; 170: 102–7.
Barrandon Y, Green H . Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A 1987; 84: 2302–6.
Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG . PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res 2008; 68: 1820–5.
Boman BM, Wicha MS, Fields JZ, Runquist OA . Symmetric division of cancer stem cells — a key mechanism in tumor growth that should be targeted in future therapeutic approaches. Clin Pharmacol Ther 2007; 81: 893–8.
Conway AE, Lindgren A, Galic Z, Pyle AD, Wu H, Zack JA, et al. A self-renewal program controls the expansion of genetically unstable cancer stem cells in pluripotent stem cell-derived tumors. Stem Cells 2009; 27: 18–28.
Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003; 113: 643–55.
Niwa H, Miyazaki J, Smith AG . Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000; 24: 372–6.
Bylund M, Andersson E, Novitch BG, Muhr J . Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci 2003; 6: 1162–8.
Hochedlinger K, Yamada Y, Beard C, Jaenisch R . Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005; 121: 465–77.
Zwaka TP, Thomson JA . Differentiation of human embryonic stem cells occurs through symmetric cell division. Stem Cells 2005; 23: 146–9.
Vogelstein B, Lane D, Levine AJ . Surfing the p53 network. Nature 2000; 408: 307–10.
Marin MC, Jost CA, Brooks LA, Irwin MS, O'Nions J, Tidy JA, et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat Genet 2000; 25: 47–54.
Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 2003; 3: 387–402.
Acknowledgements
This work was supported by a grant from the Chinese Academy of Sciences, National Natural Science Foundation of China (NSFC) 31000614 awarded to Lei DU, the “Strategic Priority Research Program” of the Chinese Academy of Sciences, Stem Cell and Regenerative Medicine Research, Grant No XDA01040409, awarded to Quan CHEN and the 973 project from the Ministry of Science and Technology of China 2009CB512800 awarded to Quan CHEN.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplementary information is available at website of Acta Pharmacologica Sinica on NPG.
Supplementary information
Supplementary Figure S1
Controls of anti-CD45, anti-CD31 and anti-CD24 antibodies. (JPG 22 kb)
Supplementary Figure S2
Immunofluorescence staining of CD44 in CD44 knockdown cells. (JPG 108 kb)
Supplementary Figure S3
Oct3/4 expression level in CD44 RNAi cells. (JPG 68 kb)
Supplementary Figure S4
Activation of Oct3/4 promoter positively correlated with Oct3/4 expression level in P6C-OPG cells. (JPG 100 kb)
Supplementary Figure S5
The proliferation of P6C cells. (JPG 69 kb)
Supplementary Figure S6
The IC50 values for the P6C cell lines at exposure of Camptothecin and 5-FU for 48 h. (JPG 83 kb)
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Rao, Gh., Liu, Hm., Li, Bw. et al. Establishment of a human colorectal cancer cell line P6C with stem cell properties and resistance to chemotherapeutic drugs. Acta Pharmacol Sin 34, 793–804 (2013). https://doi.org/10.1038/aps.2013.56
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.56
Keywords
This article is cited by
-
Targeted activation of ferroptosis in colorectal cancer via LGR4 targeting overcomes acquired drug resistance
Nature Cancer (2024)
-
Combination of gold nanoparticles with low-LET irradiation: an approach to enhance DNA DSB induction in HT29 colorectal cancer stem-like cells
Journal of Cancer Research and Clinical Oncology (2019)
-
Two novel diterpenoid heterodimers, Bisebracteolasins A and B, from Euphorbia ebracteolata Hayata, and the cancer chemotherapeutic potential of Bisebracteolasin A
Scientific Reports (2017)
-
Expression of an Oncogenic BARD1 Splice Variant Impairs Homologous Recombination and Predicts Response to PARP-1 Inhibitor Therapy in Colon Cancer
Scientific Reports (2016)
-
Cancer stem cells in human digestive tract malignancies
Tumor Biology (2016)