Abstract
A 2-μm-long Ni ion-chelated DNA molecule (Ni–DNA) was found for the first time to possess both memcapacitor and memristor properties; this Ni–DNA molecule is known as a dual memory circuit element (memelement). As a memelement, the state of impedance on Ni–DNA is proportional to the unit number of Ni ions containing a base pair complex (Ni–bp), which is determined by the previously applied external voltage. Interestingly, the impedances of Ni–DNA change in response to a change in the sweeping frequencies of the external bias. Our simulation results also indicate that changes in the effective resistance and capacitance of Ni–bp may be attributed to changes in the Ni ion redox species in the Ni–bp of a Ni–DNA nanowire. Therefore, the working mechanism of a nanowire-type memcapacitor and memristor is revealed. In summary, the Ni–DNA nanowire is shown to be a multi-dimensional memory device, whose memory state depends on the length of DNA and applied external voltages/frequencies.
Similar content being viewed by others
Introduction
Memory circuit elements (memelements) are multi-dimensional electronic devices with memory capability whose resistance, capacitance or inductance is determined by a previously applied voltage, charge, current or flux.1 Memelements have attracted a considerable amount of attention because of their potential applications in memory computing by storing and processing information on the same physical platform,2 as envisioned by Feynman in 1959.3 However, only memristor devices have been developed and characterized,4, 5 while the memcapacitor and meminductor remain to be developed, due to a lack of functional materials or systems whose capacitance or inductance can be controlled by external variables.
Several molecular electronic components,6, 7, 8 such as single-molecule electronic components9, 10 and extended metal atom chains or metal strings,6, 7, 11 have been shown to transport charges, as measured by scanning tunneling microscope (STM),3 nanopore12 and proximal probes.13 Although the electronic properties of these molecular electronic components can be controlled by external variables, they are undesirable for the development of memelements due to the instability of the scaffold of free metal atom/ion chains, the high thermionic barriers of the metal-SAM-metal structures14 and the high probability of inter-molecular interactions.
Nanowires with a high aspect ratio and large surface area-to-volume ratio have great potential for integrating with a variety of technologies, including solar cells,15, 16 fuel cells,17 integrated circuits18 and bio-sensing.19, 20 The conducting current of conventional nanowires is determined by various factors including the intrinsic composition, length, width21, 22 and chirality23 of the wires and the external bias. The impedances of conventional nanowires are invariant because they have fixed intrinsic composition, length and width. Therefore, the response current–voltage profiles of conventional nanowires monitored under direct current (DC) are unaffected by previously applied external biases. However, DNA, a long linear biopolymer formed by the linkage of four types of nucleotides, can act as a nanowire after proper doping and exhibit unique electronic properties that are not observed in conventional nanowires.18, 20
Although the DNA duplex alone exhibits poor conductance, it becomes conductive when metal ions, especially Ni ions, are chelated inside the DNA duplex (Ni–DNA).22 Recently, Ni–DNA molecules 50–100 nm in length22, 24 were shown to exhibit a negative differential resistance (NDR) effect at room temperature, a characteristic of memristive devices.21 This behavior may be attributed to the redox current (Ired) of Ni ions.24 Both chelated Ni ions and the π-stacking bases in the DNA duplex were found to be essential for charge transport in Ni–DNA nanowires.25 Therefore, it is hypothesized that the Ni ion-base chelating units (Ni–bp) in Ni–DNA may form a precise conformation that is important for charge transport.
As shown previously, the redox state of Ni ions in the Ni–DNA nanowire is switchable using the external biases.21, 24 The ratio of redox species of Ni ions (Ni2+/Ni3+) can be changed when the external biases exceed the threshold of redox potential of Ni ions,4 indicating that the Ni–DNA nanowire may exhibit a capacitive effect. In this study, a micrometer-long Ni–DNA nanowire is developed and characterized to explore its potential in the development of nanowire-based electronic devices. Interestingly, the results show that the Ni–DNA nanowire device may possess both memristor and memcapacitor functionalities. The impedance of this Ni–DNA nanowire device is tunable by external biases and bias frequencies.
Materials and methods
Ni–DNA synthesis
The pET30a vector (Novagen, Madison, WI, USA) was digested by BamHI to form a 5.4-kb linear DNA. After enzyme digestion, DNA was purified using a PCR clean-up kit (Qiagen, Valencia, CA, USA). DNA (final concentration 1.5 μM) was added to a buffered Ni2+ solution (10 mM Tris, 2.5 mM NiCl2, pH 9.0) and heated in boiling water for 5 min. After heating, the mixture was allowed to cool to room temperature overnight. The resulting Ni–DNA was dialyzed against double-deionized water to remove unbound Ni2+.
EXAFS measurement
The local structure surrounding Ni in Ni-histidine and Ni–DNA samples was explored using extended X-ray absorption fine structure (EXAFS) techniques. The Ni K-edge EXAFS measurements were performed at beamline BL07A at the Taiwan light source at the National Synchrotron Radiation Research Center, Hsinchu, Taiwan. A Si(111) Double Crystal Monochromator was used to scan the photon energy. The energy resolution (ΔE/E) for the incident X-ray photons was estimated to be 2 × 10−4. Samples were sealed between two layers of Kapton tape during measurements. The EXAFS spectra were monitored in the fluorescence mode using a Lytle fluorescence detector for both samples. To confirm the reproducibility of the experimental data, multiple scan sets were collected and compared. The IFEFFIT software package was used to analyze the EXAFS data26 to extract local structural parameters. Several possible biochemical structural models were used for fitting the experimental data with several constraints to exclude unphysical results. The coordination numbers for all neighboring shells were fixed in accordance with the adopted biochemical structural models, while the Debye–Waller factor of all shells (except for shells from bound waters) were set to be the same. The energy shifts (ΔE) that largely depend on the atomic species for O and N shells were also set to be the same, respectively.
The Ni K-edge X-ray absorption near-edge structure (XANES) spectra and EXAFS spectra of NiO, Ni2+–histidine complex, NiCl2 and Ni–DNA were collected and analyzed (Supplementary Figure S1). On the basis of the XANES and EXAFS information, the molecular conformations of Ni-base pairs (Ni–bp) were optimized using Gaussian 2009 with the basic set HF/3–21G (Figure 1b). The molecular orbitals of the HOMO and LUMO states of Ni–bps were also calculated in Gaussian 2009 (Supplementary Figure S2).
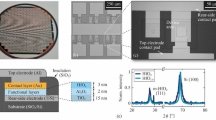
Ni ion chelating structure of Ni–DNA. (a) The χ functions (inset) and their Fourier transforms of the Ni2+–histidine complex (green) and Ni–DNA (blue) are plotted as fine and thick lines for experimental and theoretical curves, respectively. Curves are shifted vertically for clarity. (b) Configuration of the Ni–bp complex in Ni–DNA. The illustration shows that the Ni ion (deep-blue sphere) is chelated between A-T (top panel) and G-C base pairs (bottom panel). (c) AFM image of the Ni–DNA device; the blue dashed line denotes the area of two-terminal electrodes. (d) Length-dependent hysteresis I–V curves of 170 (blue), 390 (red) and 830 nm (black) long Ni–DNA devices. Experimental data and theoretical results are marked with (E) and (T), with optimal values of N0 9 × 105, 1.6 × 106, 3.01 × 106 for fitting the 170, 390 and 830 nm long Ni–DNA devices, respectively.
Substrate fabrication
Silicon wafers capped with a 300-nm thick SiO2 layer were used as substrates for device fabrication. The substrate was photo-lithographically pre-patterned to construct current leads and contact pads for wiring at the 10 and 100 μm scale, respectively. The Ti/Au electrodes with a width and thickness of ~100 and 80–100 nm, respectively, were patterned on the substrate using standard electron beam lithography to form a pair of finger electrodes with gaps from ~170 to 830 nm. Ti/Au deposition was achieved using thermal evaporation at a base pressure of ~3 × 10−6 torr for 10 min (Supplementary Figure S3).
Ni–DNA nanowire bridging
The Ni–DNA nanowires were connected between two gold terminals via DC electrophoresis to generate the Ni–DNA devices (Figure 1c and illustrated in Supplementary Figure S4). Briefly, a DC bias of 1 V is applied to the gold electrode pair in the presence of Ni–DNA (~2 μM) for 10 min, while the reference electrode is set to ground.
Electrical properties characterization
Once the Ni–DNA devices were fabricated, residual water on the devices was removed by baking the devices at 80 °C for 15 min. Electrical properties, including the current-voltage (I–V) loops and electrical writing and current reading for the memory effect, were characterized under ambient conditions with low humidity (water-free) on a Keithley 6517b instrument with a two-terminal configuration. The multiple states of the Ni–DNA memristor device were set using writing voltages of ±6 V, larger than the redox potential of Ni ions, for a certain period of time. The state of the Ni–DNA memory system was read out at reading voltages of ±1 V. For the alternating current (AC) mode reading, the current was recorded by applying a sinusoidal bias voltage across the Ni–DNA devices. The AC mode was operated at frequencies from 0.01 to 104 Hz.
The charge–voltage (q–V) measurements were also performed on the Keithley 6517b by cyclic sweeping of the bias voltage between 7 and −7 V across the Ni–DNA devices. The accumulated charge in the Ni–DNA device may be discharged during cyclic voltage sweeping, if necessary (Supplementary Figure S5).
Results and discussion
The configuration of Ni ions in a 5.4-kb linear Ni–DNA (~2.0 μm in length; see Materials and methods section) was characterized by XANES spectroscopy; the spectrum of the Ni ion in Ni–DNA was closer to that of the Ni2+–histidine complex27 than that of NiO (Supplementary Figure S1a). These results indicate that the Ni ion binds to both the oxygen and nitrogen of the bases in DNA. The shoulder near 8338 eV in the XANES spectrum of Ni–DNA (Supplementary Figure S1b) indicates that the chelated Ni ion at both redox states, Ni2+ and Ni3+, is in a five coordinate pyramidal geometry.28 The local structures surrounding the Ni ions within DNA were further refined by the distance parameters (Table 1) obtained from the extended EXAFS technique (Figure 1a). Optimal structure analysis indicates that the Ni ion is chelated between the base pairs of DNA and forms a distorted five coordinate pyramid with bound water molecules (Figure 1b), suggesting that the Ni–base pair (Ni–bp) complex is a basic conducting unit in Ni–DNA with a conformation similar to extended metal atom chains or a metal ion string.6
The charge transport properties of Ni–DNA were characterized on the two-terminal gold electrodes with various gaps (170, 390 and 830 nm). Two gold electrodes are bridged by Ni–DNA nanowires to generate a simple circuit device with the help of an external electric field. The fabricated Ni–DNA device was monitored using AFM (Figure 1c; Supplementary Figure S4). The gold electrodes of the system are indicated by blue dashed lines. The extruded area in the center of the image is the bundle of Ni–DNA nanowires with a height of ~250 nm, that is, the width of the electrode (~100 nm). The maximum number of parallel Ni–DNA nanowires in a bundle was estimated to be ~31 250. Although Ni–DNA is approximately 2 μm long, the gap width W between the paired gold electrodes was used as an effective conducting length. The simulation of the conducting currents of Ni–DNA devices indicates that the effective/conducting number of Ni–DNA nanowires in a bundle is ~1800, 1395 and 1233 for devices with a gap of 170, 390 and 830 nm, respectively (Figure 1d; Supplementary Figure S5). Interestingly, the number of Ni–DNA nanowires on Ni–DNA devices estimated by conducting current (1233–1800 nanowires) is much smaller than that (31 250 nanowires) determined by atomic force microscope (AFM). This indicates that although the thickness of a Ni–DNA bundle can be up to 250 nm, the effective conducting channel is only ~10 nm thick. The presence of an effective channel allows the same scale of conducting currents in Ni–DNA nanowires on the same gap length of terminals. Moreover, our simulation also indicates that a wider gap between electrodes leads to a lower number of effective nanowires participating in charge transport. This is consistent with the general concept of self-assembling nanowire device fabrication.29
Consistent with previous studies,21, 22, 24 the Ni–DNAs of various lengths (from 170 to 830 nm) exhibit an NDR effect (Figure 1d). The mechanism of the redox molecule-based NDR is the charge transport process of the redox reaction through the molecules between two electrodes, as reported in conventional electric chemical reactions22, 30 and in studies of the J.M. Tour group31 and our previous study.21 The bias may vary depending on the redox molecules adopted in the system. Moreover, NDR can occur in both bias directions if the appropriate redox molecule is used.32 This phenomenon was also observed in our system. In electronics, the NDR can be observed when electrons are excited with moderate electric fields to an energy and momentum beyond an inflection point in the E–k relation.33 The regular transistor-based NDR circuits are designed based on the MOSFET systems and operated in the positive range only. Accordingly, the Ni–DNA system is different from the transistor-like NDR system and may not be used for practical NDR setups, such as oscillators or filters.34
Interestingly, the peak (valley) currents of the I–V curves increase in response to the increase in the conducting length of Ni–DNA. This behavior may be attributed to the accumulative redox current (Ired) of Ni ions in the Ni–DNA bundle of the device. The redox current is proportional to the number of Ni ions (N0), which is the function of the conducting length and the number of Ni–DNA molecules in a bundle.24
To illustrate this point, we generated a theoretical model to simulate the hysteretic I–V curves of Ni–DNA using the same set of parameters, except that N0 is proportional to the length of the Ni–DNAs with the same number of Ni–DNA nanowires in a bundle. The ratio of different redox states of Ni–bp in Ni–DNA was expressed as a function of external bias (V) and time (t). The number of Ni3+-bp is denoted as N(t), serving as the state variable. N(t) was estimated using the modified Arrhenius rate equations:

where N0 is the total number of the Ni2+-bp and Ni3+-bp, kox[T,V(t)] (kred[T,V(t)]) is the oxidation (reduction) rate constant, which depends on temperature and the AC source. The kox[T,V(t)] and kred[T,V(t)] can be estimated by equation (2) and equation (3), respectively:

and

where,  and
and  are the standard rate constants of the redox reaction, which varies with length of Ni–DNA:
are the standard rate constants of the redox reaction, which varies with length of Ni–DNA:  and
and  , where mox=5.67 × 10−3, mred=6.06 × 10−7 and
, where mox=5.67 × 10−3, mred=6.06 × 10−7 and  1/s,
1/s,  1/s for 830-nm long Ni–DNA devices. Eox and Ered are the activation energies required to trigger oxidation and reduction reactions of Ni ions, respectively. Eox=3.5 eV and Ered=3.7 eV. kB is the Boltzmann constant, β=1/2 is chosen as a symmetry factor, and α=30 is adopted to tune the rate constants for a specific Ni–DNA system. The detailed model is described in the SI.
1/s for 830-nm long Ni–DNA devices. Eox and Ered are the activation energies required to trigger oxidation and reduction reactions of Ni ions, respectively. Eox=3.5 eV and Ered=3.7 eV. kB is the Boltzmann constant, β=1/2 is chosen as a symmetry factor, and α=30 is adopted to tune the rate constants for a specific Ni–DNA system. The detailed model is described in the SI.
The results of model calculations (symbols) agree well with experimental data (lines) (Figure 1d; Supplementary Figure S5). These results indicate that the redox states of Ni ions in Ni–bp can be controlled by external bias, resulting in the negative impedance effect in this molecular wire. The charge transport signal of micrometer-long Ni–DNA can be easily detected because of the redox-induced negative impedance effect. This property allows the Ni–DNA-based circuit device to fit easily into real electronic systems.
Although the chelated Ni ions are essential to charge transport in Ni–DNA nanowires, the π-stacking of bases in DNA duplexes may also play an important role.25 Molecular orbital analysis reveals that Ni ions interact with the π orbitals of base pairs (Supplementary Figure S2a) and facilitate charge transfer along the Ni–bp chain. The bound water molecules also participate in the charge transfer process (Supplementary Figure S2b), as indicated by the LUMO state of Ni–bp. During electron transport in Ni–DNA, the paired π-stacking of bp and Ni ion in Ni–bp may act as sources of electrons and holes, respectively. Therefore, the possible conducting mechanism in Ni–DNA may be the hopping of electrons from one Ni–bp unit to another. This postulation is supported by our previous findings21, 35 and the study of Wohlgamuth et al.25 The external bias-driven redox state changes of Ni ions may possibly modify the resistance and capacitance of each Ni–bp and facilitate electron transport.
The hysteretic I–V curves in Figure 1d indicate that the current flowing through the Ni–DNA system depends on the history of V(t), suggesting that the Ni–DNA system is capable of serving as a memory device. The Ni–DNA-based memory device records information via the dynamic states of Ni ions. When a constant writing voltage of 6 V—a voltage higher than that required for redox state conversion—is applied, all Ni ions in the Ni–DNA device are written to Ni3+ states, that is, N(t)/N0→1 as t→∞. These written states can be distinguished or read by the currents (read-out currents) in response to the input external voltages (written voltages). To maintain the written states of the Ni–DNA device, read-in voltages smaller than the activation energy of Ni ions were chosen in the study.
The electronic properties of the Ni–DNA system were further investigated by applying various external biases to the Ni–DNA device, and the corresponding charges were read in both DC and AC modes (Figure 2). In the DC reading mode, the Ni–DNA was first written by various external biases (±2, ±3 and ±4 V) for 5 s and the stored signal was read at a low voltage of 0.1 V. As shown in Figure 2a (symbols), different states of charge were found in Ni–DNA with different applied positive and negative writing voltages. This result shows that the state of charge in the Ni–DNA device is controlled by external voltages. Interestingly, the bias-controlled charge in the Ni–DNA system remains the same after writing. The state of charge remains stable and lasts for at least 400 s. This result clearly shows that the Ni–DNA system possesses a unique external bias- and writing time-dependent memory effect. In AC reading mode, the Ni–DNA system exhibited a sweeping speed-dependent hysteretic Q–V curve (Figure 2b). The charge in the hysteresis loops is inversely proportional to the sweeping speed.
Operation of the memcapacitor in DC mode and characteristics of hysteric Q–V curves in AC mode. (a) Reading of the Ni–DNA system (time dependence of charge accumulation, Q–t curves) of the 100 nm long Ni–DNA system at a reading voltage of 0.1 V. In the beginning, the memristor is written by applying voltages of 4, 3, 2, −2, −3 and −4 V (from top to bottom) for 5 s. Closed squares, circles and triangles indicate experimental results (E), whereas solid lines give theoretical fitting (T) to the data. (b) Hysteresis Q–V loops of the same device in (a) at different sweeping speeds. The sweeping frequencies are 3.5 × 10−2, 3.5 × 10−3 and 1.7 × 10−3 Hz for data shown as black squares, red circles and blue triangles, respectively. Solid lines denote simulation results. Symbols E and T indicate experimental data and theoretical calculations, respectively. The optimal value of N0 is 6.5 × 105.
A simulation model (Supplementary Figure S6) was also established to explain the above experimental results. The accumulated charges on the voltage-controlled Ni–DNA can be estimated by the equation: qtot(t)=qMR(t)+qMC(t), where qMR ( and qMC (qMC(t)=C(t)V(t)) denote the resistance and capacitor effects on the charge accumulation of Ni–DNA. R(t) and IMR(t) are the resistance and current flowing through the voltage-controlled Ni–DNA and can be described as
and qMC (qMC(t)=C(t)V(t)) denote the resistance and capacitor effects on the charge accumulation of Ni–DNA. R(t) and IMR(t) are the resistance and current flowing through the voltage-controlled Ni–DNA and can be described as  and
and  . However, the Ni–DNA system has an additional source of charge accumulation caused by the exchange rate of Ni ion species, or the Ni ion redox-induced effect, that is,
. However, the Ni–DNA system has an additional source of charge accumulation caused by the exchange rate of Ni ion species, or the Ni ion redox-induced effect, that is,  . Consequently, the total charge (qtot (t)) accumulation measured by the experiment can be expressed as: qtot(t)=qMR(t)+qMCt(t)+qred(t).
. Consequently, the total charge (qtot (t)) accumulation measured by the experiment can be expressed as: qtot(t)=qMR(t)+qMCt(t)+qred(t).
In DC writing mode, the contribution of qMC is ~0, and, hence, qtot(t)=qMR(t)+qred(t). Therefore, the calculated charge–time (q–t) curves at different writing voltages fit perfectly with those of experimental measurements (Figure 2a). The effective impedance values of XR of the device are determined as 6.73 × 108(Ω), 6.61 × 108(Ω) and 6.06 × 108 (Ω), when the writing biases are set at ±2, ±3 and ±4 V, respectively. The effective capacitances of the device are 3.14 × 10−10(F), 3.08 × 10−10(F) and 2.73 × 10−10(F) at writing biases of ±2, ±3 and ±4 V, respectively.
Interestingly, in AC writing mode, the calculated charge–voltage (q–V) sweeping curves fit well with the experimental results (Figure 2b). This observation indicates that the contribution of qMR can be neglected, and, hence, qtot is controlled by the voltage sweeping frequency. The qred term dominates qtot at low sweeping frequency; by contrast, qMC dominates qtot at high sweeping frequency. The effective impedance XR and capacitance of the Ni–DNA device under sweeping frequencies of 3.5 × 10−2, 3.5 × 10−3 and 1.7 × 10−3 Hz were determined to be in the range of 6 × 108–7.2 × 108 (Ω) and 2.7 × 10−10–3.5 × 10−10 (F), respectively. The results from model calculations (dashed lines in Figure 2) agree well with experimental measurements (solid lines in Figure 2), suggesting that the Ni–DNA system is not only a memristor but also a voltage-induced memcapacitive system.35
The effect of different read-in voltage waveforms, for example, sinusoidal and square waveforms, on the read-out current of the Ni–DNA memory device was studied. After writing at 6 V for a half-period with a sinusoidal or square waveform, the read-out currents were recorded as shown in Figure 3a. The read-out current is stable when the sinusoidal read-in voltage waveform is applied. However, when operated with the square read-in waveform, the read-out currents exhibit a large high-frequency oscillation at the point where the abrupt jump occurs. The high-frequency oscillation in the read-out current is caused by parasitic capacitance. When the square read-in waveform is applied, a singularity  occurs at the jump discontinuities V(t). The portion of the current through the memcapacitor can be expressed as
occurs at the jump discontinuities V(t). The portion of the current through the memcapacitor can be expressed as  . Singular behavior occurs at the jump discontinuities V(t) because of the term
. Singular behavior occurs at the jump discontinuities V(t) because of the term  . Compared to the unsmooth read-in voltage waveform, the smooth read-in voltage waveform (for example, the sine read-in) is more suitable as the input read-in voltage for Ni–DNA-based memory devices.
. Compared to the unsmooth read-in voltage waveform, the smooth read-in voltage waveform (for example, the sine read-in) is more suitable as the input read-in voltage for Ni–DNA-based memory devices.
Applications of memristor functionality: a memory device in AC mode. (a) Comparison between the read-out current in R operation caused by square (black empty square and line) and sinusoidal (red empty square and line) read-in voltages. Inset shows the high-frequency oscillation in the read-out current caused by jump discontinuity in the square read-in voltage. (b) (upper panel) Read-in voltage for a series of write–read–erase operations. (middle panel) N(t)/N0 ratios in the series of write (W+ and W−)–read (R)–erase (E) operations. (lower panel) Read-out currents as a function of sine read-in voltage in R operations immediately after W+, E and W− operations. (c) I–V curves of the Ni–DNA device after applying three different writing conditions. (E) and (T) denote experimental data and theoretical calculation results, respectively. (d) Comparison among net read-out currents in continuous and pulsed reading modes. Red and black open squares correspond to continuous and pulsed reading modes, respectively. Solid lines represent theoretical calculation results.
The Ni–DNA system as a memory device was further investigated by operations of positive writing (W+), reading (R), erasing (E) and negative writing (W−) (Figure 3b, upper panel). The W+ (W−) operation was performed by applying a constant input voltage at 6 V (−6 V) across the electrodes for 12 s. The magnitude of the input voltages in the W+ (W−) operation was set higher than the activation energy Eox (Ered) of the oxidation (reduction) reaction of Ni ions. Consequently, the state of Ni ions near 100% (or 0%) Ni3+ via oxidation (or reduction) in the Ni–DNA memory system was recorded (Figure 3b, middle panel). The E operation was performed by applying a high-frequency dampened sinusoidal voltage with a peak of 1.5 V. The R operation was performed by reading the output currents corresponding to the sinusoidal input voltage at 1.5 V with a period of 16 s. Experimental measurements and model calculations were performed to obtain read-out currents from the R operations. The read-out currents in the R operation agree well with the experiment and theory. As shown in Figure 3b (lower panel), the state of Ni ions recorded by the W+ or W− operation can be read out by the R operation. The read-out current in the R operation immediately after W+ can be readily distinguished from that in the R operation immediately after W−. By contrast, the read-out currents in the R operations immediately after E operation are indistinguishable from those of R operations immediately after either W+ or W−. Information on the state of Ni ions could be erased by the E operation, resulting in an abrupt reduction of N(t)/N0 from the initial state to 42.4%, which is a chemical equilibrium value in the closed circuit under an ambient environment (Supplementary Figure S7). The writing and reading processes are reversible. Over 700 writing/reading cycles could be performed on the fabricated Ni–DNA devices without losing much of the signal. All control data are presented in the Supplementary Section and Supplementary Figures S8 and S9.
According to the phase diagram shown in Figure 3c, the Ni–DNA nanowire is postulated to operate as a multi-state memory device when the redox states of a single Ni ion can be finely tuned by the writing voltage. The multi-state memory can be quantified via the value  of the system after the W+ operation (Figure 3d). Here, IP is the peak value of the read-out current and IV is the valley value of the read-out current. Both continuous and pulsed reading modes were used to check the retention abilities of the Ni–DNA memory system. Interestingly, the memory information in the Ni–DNA system can be maintained for as long as 19 h without being erased by repeated W+–E–W−–E operation cycles. By contrast, the memory information can only be retained for ~30 min in the continuous reading mode. This result indicates that the reading operation may affect the stability of the multi-state memory system. A similar result was also observed in the theoretical model mentioned in the SI section. However, the extraordinarily high retention ability of Ni–DNA in the pulsed reading mode contradicts the theoretical modeling and can be attributed to the nanoscale contacts between Ni–DNA and metal electrodes. The nanoscale contacts build a barrier, working as parasitic capacitance to confine carriers in the Ni–DNA system.
of the system after the W+ operation (Figure 3d). Here, IP is the peak value of the read-out current and IV is the valley value of the read-out current. Both continuous and pulsed reading modes were used to check the retention abilities of the Ni–DNA memory system. Interestingly, the memory information in the Ni–DNA system can be maintained for as long as 19 h without being erased by repeated W+–E–W−–E operation cycles. By contrast, the memory information can only be retained for ~30 min in the continuous reading mode. This result indicates that the reading operation may affect the stability of the multi-state memory system. A similar result was also observed in the theoretical model mentioned in the SI section. However, the extraordinarily high retention ability of Ni–DNA in the pulsed reading mode contradicts the theoretical modeling and can be attributed to the nanoscale contacts between Ni–DNA and metal electrodes. The nanoscale contacts build a barrier, working as parasitic capacitance to confine carriers in the Ni–DNA system.
Recently, a new computer paradigm known as a ‘memcomputer’ was proposed to tackle notoriously difficult problems by bypassing the limits of conventional computers. The brain-like configuration alternative to quantum computing employs memory elements to process and store information at the same physical location.2, 36 This universal memcomputing machine is able to solve nondeterministic polynomial time complete problems in polynomial time.37 The basic building blocks in the circuit design of a memcomputer are memristors, memcapacitors and/or meminductors.2 Interestingly, the impedance of the developed Ni–DNA device was demonstrated to be tunable by external bias, writing time and writing frequency, making it an ideal two-dimensional memory and operating system with both memristor and memcapacitor functionalities. Moreover, its resistance is independent of the capacitance. The function of Ni–DNA impedance states can be used simultaneously for calculation and information storage, a unique characteristic of memcomputing. Thus, the fabrication of a memcomputing device can possibly be realized by integrating Ni–DNA-based devices.
Conclusions
On the basis of the principles of DNA origami and self-assembly, DNA-guided nickel ion molecular wires can be fabricated using green, environmentally friendly methods. The Ni–DNA system developed here was shown to be a memcomputing device, exhibiting both memcapacitor and memristor functionalities and the NDR effect (Supplementary Figure S5). The device is controllable using external bias, writing time and writing frequencies. This Ni ion-containing DNA is thus a promising one-dimensional nanomaterial useful as a scalable template for molecular electronic devices and computing machines.
References
Pershin, Y. V. & Di Ventra, M. Memcomputing: A Computing Paradigm to Store and Process Information on the Same Physical Platform, 2014 International Workshop on Computational Electronics (IWCE): Paris, France (2014).
Di Ventra, M. & Pershin, Y. V. The parallel approach. Nat. Phys. 9, 200–202 (2013).
Lafferentz, L., Ample, F., Yu, H., Hecht, S., Joachim, C. & Grill, L. Conductance of a single conjugated polymer as a continuous function of its length. Science 323, 1193–1197 (2009).
Chu, H. L., Chiu, S. C., Sung, C. F., Tseng, W., Chang, Y. C., Jian, W. B., Chen, Y. C., Yuan, C. J., Li, H. Y., Gu, F. X., Di Ventra, M. & Chang, C. C. Programmable redox state of the nickel ion chain in DNA. Nano Lett. 14, 1026–1031 (2014).
Strukov, D. B., Snider, G. S., Stewart, D. R. & Williams, R. S. The missing memristor found. Nature 453, 80–83 (2008).
Liu, I. P. C., Wang, W. Z. & Peng, S. M. New generation of metal string complexes: strengthening metal-metal interaction via naphthyridyl group modulated oligo-alpha-pyridylamido ligands. Chem. Commun. 2009, 4323–4331 (2009).
Berry, J. F., Cotton, F. A., Lei, P., Lu, T. B. & Murillo, C. A. Additional steps toward molecular scale wires: further study of Ni-5(10/11+) chains embraced by polypyridylamide ligands. Inorg. Chem. 42, 3534–3539 (2003).
Berry, J. F., Cotton, F. A., Lei, P. & Murillo, C. A. Further structural and magnetic studies of tricopper dipyridylamido complexes. Inorg. Chem. 42, 377–382 (2003).
Bumm, L. A., Arnold, J. J., Cygan, M. T., Dunbar, T. D., Burgin, T. P., Jones, L., Allara, D. L., Tour, J. M. & Weiss, P. S. Are single molecular wires conducting? Science 271, 1705–1707 (1996).
Joachim, C., Gimzewski, J. K., Schlittler, R. R. & Chavy, C. Electronic transparency of a single C-60 molecule. Phys. Rev. Lett. 74, 2102–2105 (1995).
Roth, S. One-Dimensional Metals: Physics and Materials Science, 1st edn (Wiley: New York, NY, USA, 1995).
Akkerman, H. B., Blom, P. W. M., de Leeuw, D. M. & de Boer, B. Towards molecular electronics with large-area molecular junctions. Nature 441, 69–72 (2006).
Davis, J. J., Morgan, D. A., Wrathmell, C. L., Axford, D. N., Zhao, J. & Wang, N. Molecular bioelectronics. J. Mater. Chem. 15, 2160–2174 (2005).
Chen, J., Wang, W., Klemic, J., Reed, M. A., Axelrod, B. W., Kaschak, D. M., Rawlett, A. M., Price, D. W., Dirk, S. M., Tour, J. M., Grubisha, D. S. & Bennett, D. W. Molecular wires, switches, and memories. Ann. NY Acad. Sci 960, 69–99 (2002).
Garnett, E. C., Brongersma, M. L., Cui, Y. & McGehee, M. D. Nanowire solar cells. Annu. Rev. Mater. Res. 41, 269–295 (2011).
Liu, X., He, L. C., Zheng, J. Z., Guo, J., Bi, F., Ma, X., Zhao, K., Liu, Y. L., Song, R. & Tang, Z. Y. Solar-light-driven renewable butanol separation by core-shell Ag@ZIF-8 nanowires. Adv. Mater. 27, 3273–3277 (2015).
Yin, H., Zhao, S., Zhao, K., Muqsit, A., Tang, H., Chang, L., Zhao, H., Gao, Y. & Tang, Z. Ultrathin platinum nanowires grown on single-layered nickel hydroxide with high hydrogen evolution activity. Nat. Commun 6, 6430 (2015).
Friedman, R. S., McAlpine, M. C., Ricketts, D. S., Ham, D. & Lieber, C. M. Nanotechnology high-speed integrated nanowire circuits. Nature 434, 1085–1085 (2005).
Li, Z., Zhu, Z., Liu, W., Zhou, Y., Han, B., Gao, Y. & Tang, Z. Reversible plasmonic circular dichroism of Au nanorod and DNA assemblies. J. Am. Chem. Soc. 134, 3322–3325 (2012).
Hahm, J.-I. & Lieber, C. M. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 4, 51–54 (2004).
Jangjian, P.-C., Liu, T.-F., Li, M.-Y., Tsai, M.-S. & Chang, C.-C. Room temperature negative differential resistance in DNA-based molecular devices. Appl. Phys. Lett. 94, 043105 (2009).
Jang Jian, P.-C., Liu, T.-F., Tsai, C.-M., Tsai, M.-S. & Chang, C.-C. Ni2+ doping DNA: a semiconducting biopolymer. Nanotechnology 19, 355703 (2008).
Zheng, J. Z., Wu, Y. J., Deng, K., He, M., He, L. C., Cao, J., Zhang, X. G., Liu, Y. L., Li, S. X. & Tang, Z. Y. Chirality-discriminated conductivity of metal-amino acid biocoordination polymer nanowires. ACS Nano 10, 8564–8570 (2016).
Tseng, S. H., JangJian, P. C., Tsai, C. M., Cheng, T. M., Chu, H. L., Chang, Y. C., Chung, W. H. & Chang, C. C. Ni2+-Enhanced charge transport via pi-pi stacking corridor in metallic DNA. Biophys. J. 100, 1042–1048 (2011).
Wohlgamuth, C. H., McWilliams, M. A. & Slinker, J. D. DNA as a molecular wire: distance and sequence dependence. Anal. Chem. 85, 8634–8640 (2013).
Ravel, B. & Newville, M. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12, 537–541 (2005).
Gesawat, A. A., Ahmed, M. A. & Shakeel, F. Solution study of ternary complexes of nickel(Ii) containing histidine and diols. J. Chil. Chem. Soc. 55, 304–306 (2010).
Colpas, G. J., Maroney, M. J., Bagyinka, C., Kumar, M., Willis, W. S., Suib, S. L., Baidya, N. & Mascharak, P. K. X-Ray spectroscopic studies of nickel-complexes, with application to the structure of nickel sites in hydrogenases. Inorg. Chem. 30, 920–928 (1991).
Li, Z. T., Zhu, Z. N., Liu, W. J., Zhou, Y. L., Han, B., Gao, Y. & Tang, Z. Y. Reversible plasmonic circular dichroism of Au nanorod and DNA assemblies. J. Am. Chem. Soc. 134, 3322–3325 (2012).
Jangjian, P. C., Liu, T. F., Tsai, C. M., Li, M. Y., Tsai, M. S., Tseng, S. H., Cheng, T. M. & Chang, C. C. DNA mismatch detection by metal ion enhanced impedance analysis. Chin. J. Phys. 47, 740–747 (2009).
Chen, J., Reed, M. A., Rawlett, A. M. & Tour, J. M. Large on-off ratios and negative differential resistance in a molecular electronic device. Science 286, 1550–1552 (1999).
Xue, Y. Q., Datta, S., Hong, S., Reifenberger, R., Henderson, J. I. & Kubiak, C. P. Negative differential resistance in the scanning-tunneling spectroscopy of organic molecules. Phys. Rev. B 59, R7852–R7855 (1999).
Esaki, L. & Tsu, R. Superlattice and negative differential conductivity in semiconductors. IBM J. Res. Dev. 14, 61–65 (1970).
Chen, S. L., Griffin, P. B. & Plummer, J. D. Negative differential resistance circuit design and memory applications. IEEE Trans. Electron Dev. 56, 634–640 (2009).
Di Ventra, M., Pershin, Y. V. & Chua, L. O. Circuit elements with memory: memristors, memcapacitors, and meminductors. Proc. IEEE 97, 1717–1724 (2009).
Traversa, F. L. & Di Ventra, M. Universal memcomputing machines. IEEE Trans. Neural Netw. Learn. Syst. 26, 2702–2715 (2015).
Traversa, F. L., Ramella, C., Bonani, F. & Di Ventra, M. Memcomputing NP-complete problems in polynomial time using polynomial resources and collective states. Sci. Adv. 1, e1500031 (2015).
Acknowledgements
We thank Prof. PC Huang and Prof. T Randall Lee for valuable comments and discussion. This study is supported in part by the Ministry of Science and Technology (MOST), Taiwan (ROC) MOST 103-2112-M-009-011-MY3, MOST 104-2627-M-009-007, MOST 105-2627-M-009-006 (C-CC) and MOST 103- 2112-M-009-014-MY3 (Y-CC). This work is partially supported by “Aiming for the Top University Program” of the National Chiao Tung University and Ministry of Education, Taiwan (ROC). We thank the National Center for Theoretical Sciences, the National Synchrotron Radiation Research Center for the use of XANES and EXAFS and the National Center for High-performance Computing for computing time and facilities. Ms Ya-Hui Lin is acknowledged for preparing the DNA samples used in this study.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims inpublished maps and institutional affiliations.
Supplementary Information accompanies the paper on the NPG Asia Materials website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chu, HL., Lai, JJ., Wu, LY. et al. Exploration and characterization of the memcapacitor and memristor properties of Ni–DNA nanowire devices. NPG Asia Mater 9, e430 (2017). https://doi.org/10.1038/am.2017.157
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/am.2017.157
This article is cited by
-
Dynamical nonlinear memory capacitance in biomimetic membranes
Nature Communications (2019)
-
The fabrication and application of Ni-DNA nanowire-based nanoelectronic devices
Nano Research (2019)